Original Article - DOI:10.33594/000000679
Accepted 15 December 2023 - Published online 8 January 2024
RING1 Inhibition Has a Cell-Specific Antitumoral Role by Promoting Autophagy in Endometrial Cancer Cells
Keywords
Abstract
Background/Aims:
Factors influencing gene expression through chemical modifications of histones may play an important role in the regulation of the autophagy process in cancers. RING1A or RING1B are responsible for the catalytical activity of Polycomb repressive complex 1 (PRC1) which monoubiquitylate histone H2A. The aim of the study was to determine the effect of the RING1A/B protein inhibition on the autophagy process in endometrial cancer cells and the anticancer effectiveness of RING1 inhibitor PRT4165 in combination with autophagy inhibitors.Methods:
The expression of autophagy genes and proteins were analyzed in endometrial cancer cells HEC-1A and Ishikawa grown in different glucose concentrations and treated with PRT4165. To assess the effectiveness of PRT4165 used alone or in combination with HCQ or Lys05, IC50 and the combination index (CI) were calculated. Flow cytometry method was used to estimate apoptotic cells after treatment.Results:
The results confirm the impact of RINGs on autophagy and apoptosis in endometrial cancer cells. PRT4165 inhibitor causes changes in the expression of ATG genes and autophagy markers and the effect depends on glucose concentration and cell types. However, the anticancer effectiveness of PRT4165 was lower when it was used in combination with autophagy inhibitors, suggesting that such a combination is not a promising anticancer strategy.Conclusion:
The results indicate the importance of the RINGs in the process of autophagy and apoptosis. Further potentially more effective combinations of PRT4165 with autophagy modulators should be sought.Introduction
Endometrial cancer is the sixth most common cancer in women. Although it is usually diagnosed during and after menopause, an increase in the number of younger women affected is observed [1, 2]. Due to the insufficient clinical effectiveness of the chemotherapies used in the treatment of endometrial cancer, new therapeutic strategies aimed at cellular processes related to cell survival and resistance to therapy are sought [3]. One such process is autophagy, whose dysregulation can play an important role in cancer, although its function in the carcinogenesis process remains ambiguous. Autophagy, which contributes to the removal of redundant, damaged, and harmful cellular elements, is believed to prevent cancer formation. However, in advanced cancers exposed to nutrient deficiency, autophagy promotes their survival and growth [4, 5]. This two-faced role of autophagy is well-seen in the case of endometrial cancer [6]. Metformin which enhances autophagy via AMPK-mediated mTOR inactivation, has been reported to reduce the risk of endometrial cancer. Thus, autophagy inducers may be useful for chemoprevention of endometrial cancer. In contrast, autophagy appears to promote endometrial cancer once it is established [6]. Induction of autophagy depends on the availability of nutrients (glucose, amino acids) and the presence of insulin or growth factors [7]. Regulation of autophagy is related to ATG genes coding for proteins that are involved in particular stages of the process. During autophagy, the created phagophore engulfs cytosolic components including organelles, and closes, forming an autophagosome, which subsequently fuses with lysosome, leading to the proteolytic degradation of the cargo [8, 9]. Essential for the elongation of phagophore are two ubiquitin-like conjugation mechanisms involving Atg12-Atg5-Atg16L complex and cleavage of pro-LC3 by the protease Atg4 to create LC3-I (LC3A), which is transformed to LC3-II (LC3B) by conjugation to phosphatidylethanolamine on the growing phagophore membrane. The relative expression of LC3-I and LC3-II are considered to be autophagy markers [8-10]. The other marker of autophagy - p62, is a ubiquitin- and LC3-binding protein. As autophagy is activated, p62 binds to the ubiquitinated cargoes and delivers them to the autophagosome for degradation where it is itself degraded along with the cargoes. Recent studies indicate that epigenetic factors influencing gene expression through chemical modifications of histones may play an important role in the regulation of the autophagy process [11]. Polycomb repressive complex 1 responsible for the monoubiquitinylation of histone H2A lysine 119 is one of the main regulators of gene expression [12]. BMI-1 is the best-studied component of the PRC1 complex and plays a vital role in many cellular processes, including cell proliferation, immortalization, and aging [13]. Recent studies suggest that the BMI-1 protein may be involved in the inhibition of autophagy in some types of cancer [14]. The importance of the other components of the PRC1 complex for tumor development and progression is under investigated. RING1A and RING1B proteins are constituents of PRC1 and are responsible for the enzymatic activity of the complex [12]. There is not much data concerning the role of RINGs in the autophagy process. RING1A/B activity has been linked with autophagy only via AMBRA1 protein. AMBRA1 is responsible for the promotion of autophagy by activation and stabilization of the Beclin 1-Vps34 complex [15]. It was shown that RING1B caused ubiquitination of AMBRA1 and as a result, reduction of autophagy [16].
Our study aimed to determine the effect of the RING1A and RING1B protein inhibition on the autophagy process in endometrial cancer cells and the anticancer effectiveness of RING1 inhibitor PRT4165 in combination with autophagy inhibitors. The results of the studies indicate the importance of the RINGs proteins in the process of autophagy and apoptosis in endometrial cancer cells, but their influence depends on the type of cells.
Materials and Methods
Cell culture and treatment
Experiments were conducted on endometrial cancer cells HEC-1A (American Type Culture Collection, Manassas, VA) and Ishikawa (European Collection of Authenticated Cell Cultures, Wiltshire, UK). Both endometrial cancer cell lines were grown in DMEM: F12 media (BioWest, France) containing 10% (HEC-1A) or 5% (Ishikawa) (v/v) FBS under standard conditions (37°C, 5% CO2).
HEC-1A and Ishikawa cells were treated with PRT4165 (MedChemTronica, USA) at concentrations corresponding to IC25 or IC50 values in standard culture conditions or low glucose concentration (0, 5 mM). The effect of treatment was checked after 48 hours. Knockdown experiments were performed using Silencer Select siRNA (ID: s12035, s12069 ) (Ambion®, Carlsbad, CA, USA). To knockdown RING1 and RNF2 siRNA targeting both genes were complexed to Lipofectamine RNAiMAX (InvitrogenTM, ThermoFisher Scientific, Grand Island, NY, USA) following the manufacturer’s specifications. The siRNAs were used at a concentration of 30 nM. All treatments were performed at least in triplicate and three independent experiments were carried out.
RNA isolation, cDNA synthesis and RT-PCR
Total RNA from cells was isolated using the Tissue Total RNA GPB Mini Kit (Genoplast Biochemicals, Poland) following the manufacturer’s protocol and quantified spectrophotometrically. After RNA isolation reverse transcription reaction was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystem, USA), according to the included protocol. PCR reactions were performed using the Mastercycler ep realplex (Eppendorf, Hamburg, Germany). Specific sequences of primers for RING1, RNF2, MAP1LC3, SQSTM1, BECN1, ATG3, ATG5, ATG7, ATG12 and HPRT1 are in Supplementary Table 1. The equation 1000*2-ΔCt was applied to calculate the expression of studied genes, where ΔCt = Ct of the target gene – Ct the reference gene (HPRT1). Results are expressed as a number of target gene mRNA copies per 1000 copies of HPRT1 mRNA. The fold differences in genes expression in cells normalized to HPRT1 levels were calculated using the formula 2ΔΔCt.
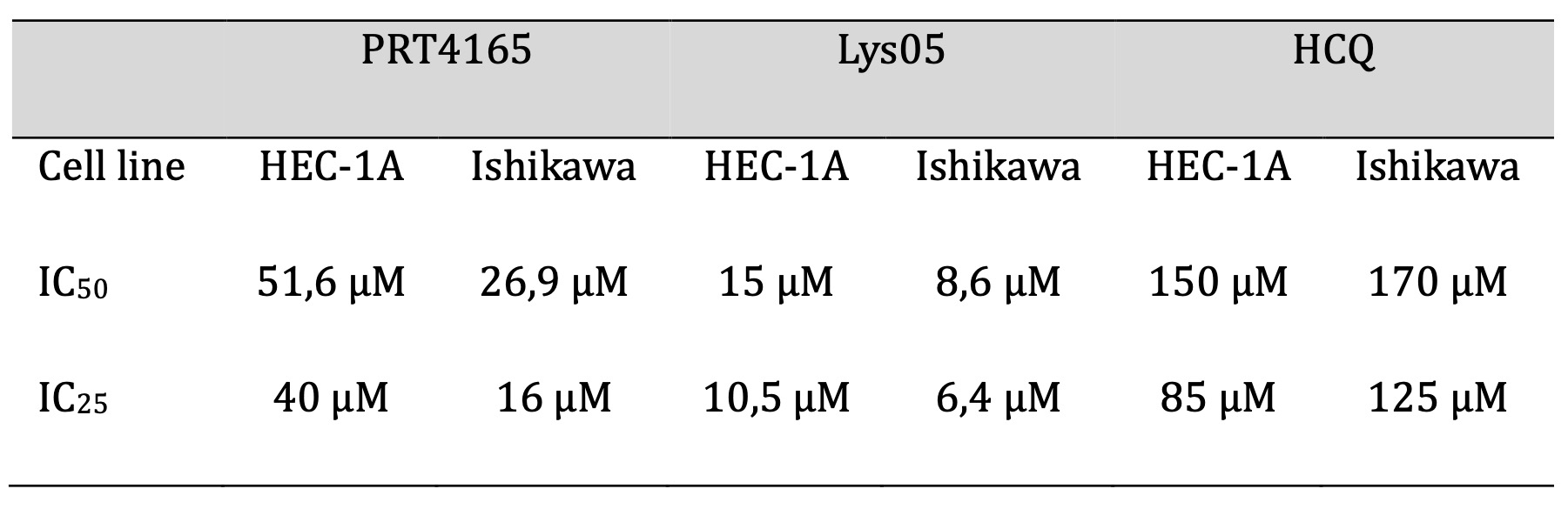
Table 1: IC50 and IC25 values were calculated for PRT4165, Lys05, and HCQ in HEC-1A and Ishikawa cell lines
Western Blotting
Endometrial cancer cells were lysed in a RIPA buffer (50 mM Tris HCl pH 8, 8; 150 mM NaCl, 1% Nonidet P-40, 0, 5% sodium deoxycholate, 0, 1% SDS, 1 mM EDTA, 1 mM PMSF). Concentrations of protein were determined using the Lowry method. Proteins of the cell lysates were resolved by 10%, 12, 5% and 15% SDS-PAGE and transferred to Immobilon P membranes. The blots were incubated overnight at 4°C with the following primary antibodies: anti-RING1A (#13069, Cell Signaling Technology, USA), anti-RING1B (#5694, CST, USA), anti-LC3 (#12741, CST, USA), anti-p62 (#5114, CST, USA), anti-H2AK119Ub (#8240, CST, USA), anti-phospho-RIP (#65746, CST, USA), anti-RIP (#3493, CST, USA), anti-MLKL (#14993, CST, USA), anti-β-actin (sc-47778, Santa Cruz Biotechnology, Dallas, TX, USA). Goat anti-mouse and anti-rabbit secondary antibodies conjugated with horseradish peroxidase (CST, USA) were used as secondary antibodies. The bands visualized by immunodetection were analyzed by densitometry using a Gel-Pro Analyzer software version 3.0 (Media Cybernetics, Inc., Bethesda, MD, USA). The IOD (integrated optical density) obtained in the Gel Pro program was used to estimate relative protein expression. The relative protein level is presented as a ratio of the IOD of the bands corresponding to the analyzed protein in each sample and the IOD of B-actin in the same sample.
MTT assay
The MTT assay was used to determine the viability of cells treated with compounds. Cells were seeded in 96-well plates at a density of 10 x 103 cells per well (HEC-1A) or 8 x 103 per well (Ishikawa). After 24 hours, cells were treated with PRT4165, autophagy inhibitors – Lys05 and HCQ, combinations of compounds, and then incubated for 48 hours. 20 μl of tetrazole salt solution was added to each well and the plate was placed in an incubator for 4 hours. After incubation, the medium was removed and 50 μl of dimethyl sulfoxide was added to each well to dissolve the formed formazan crystals. The absorbance was measured at a wavelength of 570 nm. The reagent control absorbance was subtracted from the single result. The percentage of viable cells was calculated by comparing the absorbance values of the test samples with the absorbance values of the control samples (untreated cells). The control absorbance was taken as 1.
Combination Index
Using the Chou-Talalay method, the CI (Combination Index) was calculated, the value of which determines how the compounds interact in the combinations used. CI < 0.9 is assumed as a synergistic effect, CI = 0.9 – 1.1 as an additive effect, and CI > 1.1 as an antagonistic effect [17]. To determine the type of interactions between the compounds in the combinations used, CI calculations were performed using the obtained IC values.
Flow cytometry
Cell survival in control and test samples was determined by flow cytometry using the Vybrant Apoptosis Assay Kit #4. The principle of the test is based on the cytometric evaluation of the penetration of the green dye (YO-PRO-1) into apoptotic cells and the second fluorescent dye, propidium iodide (PI), which penetrates mainly into dead cells and cells undergoing apoptosis later.
After incubation, cells were harvested and centrifuged at 300 rpm for 5 minutes at room temperature. The obtained pellet was resuspended in 0, 5 ml of PBS with the addition of both dyes (0, 5 μl of each dye). The samples thus obtained were incubated for 15 minutes in the dark, followed by a cytometric measurement.
Statistical analysis
The statistical analyses were performed using a GraphPad Prism 8.0 program (Graph-Pad Sotfware Inc., San Diego, CA, USA). The Student’s unpaired t-test was used to compare the differences between treated and control cells. Groups were analyzed using the nonparametric Kruskal-Wallis test with the post-hoc Dunn test. A p-value < 0, 05 was considered to indicate a statistically significant difference. depends on the type of cells.
Results
Effect of RINGs on the expressions of genes involved in autophagy.
The RNA interference method was used to silence the expression of RING1 and RNF2 (coding for RING1A and RING1B, respectively) in HEC-1A and Ishikawa cells. In cells treated with specific siRNAs, RING1 and RNF2 expressions were substantially reduced (Fig. 1). RING1 silencing caused decreased expression of MAP1LC3 and increased expression of SQSTM1 in both cell lines. These genes encode for LC3 and p62 proteins, which are considered as autophagy markers. Expression of the other autophagy genes did not change in HEC-1A cells. In Ishikawa cells, the mRNA expression levels of BECN1, ATG3, ATG7, and ATG12 were lower in cells with RING1A depletion. Down-regulation of RNF2 caused increased expression of MAP1LC3 and LC3B in both cells. Moreover, in Ishikawa cells, the increased expression of BECN1, ATG3, and ATG12 was observed. Interestingly, in both cells, downregulation of RING1 caused increased expression of RNF2, but not vice versa.
In mammalian cells, the regulation of autophagy by amino acids, and also by the hormone insulin, has been extensively investigated, but knowledge about the effects of other autophagy regulators, including another nutrient, glucose, is more limited. Thus, we checked the effect of hypoglycemia on RINGs and autophagy gene expression. The expression of RING1A/B was dependent on glucose. In both cell types growing in medium with low glucose mRNA level of RING1 is higher compared to levels in cells growing in high glucose. In contrast, the protein level of RING1A protein is lower in low glucose in both cell types. Low glucose caused also an increased mRNA of RING1B and a decreased level of protein, but only in Ishikawa cells. This may suggest that increased mRNA level is for compensation of higher turnover of RINGs (Fig. 2). Low glucose caused also increased levels of LC3 and p62 proteins and that was correlated with their mRNA expression increase. Moreover, in cells growing in low glucose, the expression of BECN1 and ATG12 was higher than in cells growing in 25 mM glucose (Supplementary Fig. 1). These results confirm that glucose deprivation activates autophagy in endometrial cancer cells.
Next the PRT4165 (ang. 2-(3-pyridinylmethylene)-1H-indene-1, 3(2H)-dione) was used to inhibit the activity of RING1A/B in cells growing in low and high glucose. PRT4165 was used in a concentration of IC50, that was previously determined by MTT assay. The results show that PRT4165 treatment differently affects mRNA and protein levels dependent on glucose concentration, which is especially seen in Ishikawa cells (Fig. 3A, B). In cells treated with PRT4165 and grown in high glucose lower RNF2 expression was correlated with higher LC3B protein level and lower p62 protein level, which suggests the activation of autophagy (Fig. 3D, F). This effect is not seen in low glucose since the PRT4165 treatment caused an increase of RNF2 expression and the amount of LC3B did not increase (Fig. 3D, F). The level of p62 is lower but this was associated with a lower transcript amount. In HEC-1A cells PRT4165 treatment caused in both glucose concentrations a higher amount of p62, which may suggest the inhibition of autophagy (Fig. 3C, E). The PRT4165 treatment caused the decrease of histone H2A ubiquitination level on lysine 119 in both cell types in low glucose. In the case of cells growing in high glucose the reduced level was seen mostly in Ishikawa cells (Fig. 4).
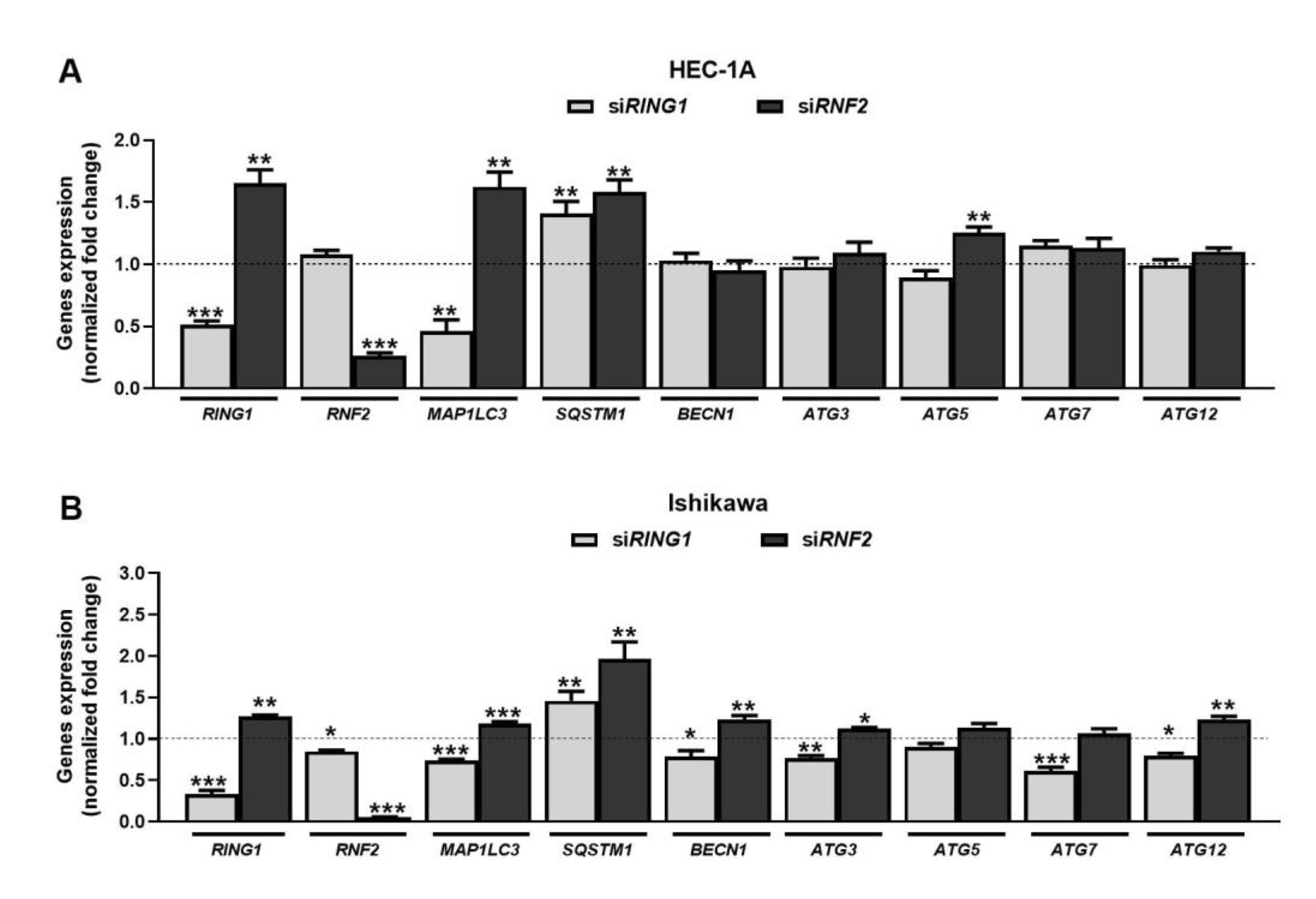
Fig. 1: Effect of RING1/RNF2 downregulation on expression of genes involved in autophagy in HEC-1A (A) and Ishikawa (B) cells. Cells were treated with 30 nmol/L RING1/RNF2 siRNA or scrambled siRNA (control). Changes in RING1, RNF2, MAP1LC3, SQSTM1, BECN1, ATG3, ATG5, ATG7, and ATG12 mRNA levels were analyzed using the Real-Time PCR method. All experiments were performed in triplicate and the obtained results are shown as means ± SEM; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
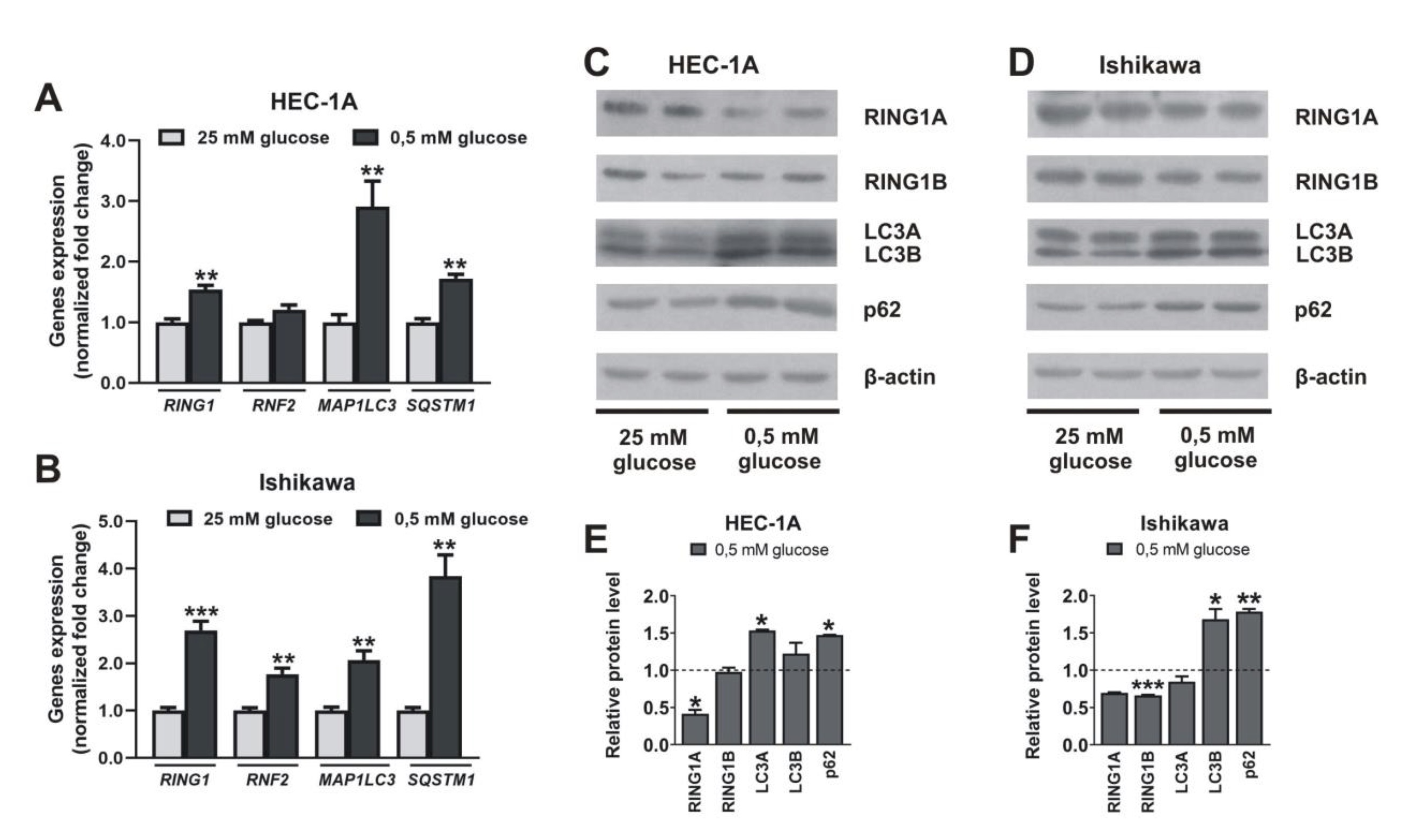
Fig. 2: Impact of hypoglycemia on RING1A/B, LC3, and p62 expression. HEC-1A and Ishikawa cells were incubated in standard conditions (25 mM glucose) and hypoglycemia (0,5 mM glucose) for 48 h. Changes in RING1, RNF2, MAP1LC3, and SQSTM1 mRNA levels were analyzed using the Real-Time PCR method (A, B). RING1A/B pro-tein levels as well as autophagy markers LC3A/B and p62 were analyzed by Western blot (C, D). The intensity of bands corresponding to proteins after Western blot was analyzed by densitometry. The results are shown as the fold change of proteins levels of treated cells vs. control (untreated cells) in HEC-1A (E) and Ishikawa (F) cells. All experiments were performed in triplicate and the obtained results are shown as means ± SEM; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
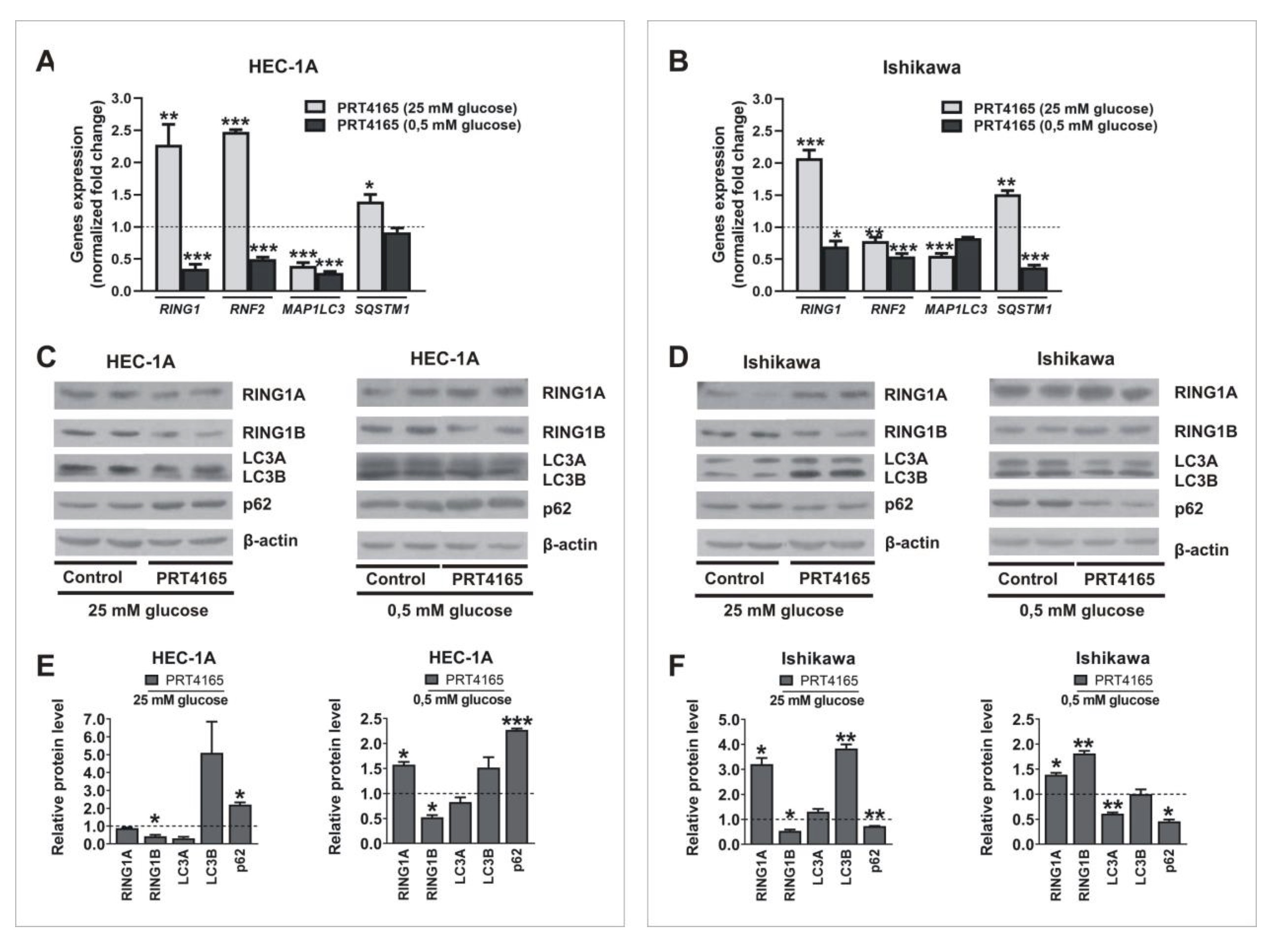
Fig. 3: Impact of PRT4165 on RING1A/B, LC3 and p62 expression in different glucose concentrations. HEC-1A and Ishikawa cells were treated with PRT4165 (51,6 µM and 26,9 µM respectively) in standard conditions and hypoglycemia for 48 h. Changes in RING1, RNF2, MAP1LC3 and SQSTM1 mRNA levels in cells treated with PRT4165 were analyzed using the Real-Time PCR method (A, B). RING1A/B protein levels as well as autophagy markers LC3A/B and p62 were analyzed by Western blot (C, D). The intensity of bands corresponding to proteins after Western blot was analyzed by densitometry. The results are shown as the fold change of proteins levels of treated cells vs. control (untreated cells) in HEC-1A (E) and Ishikawa (F) cells. All experiments were performed in triplicate and the obtained results are shown as means ± SEM; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
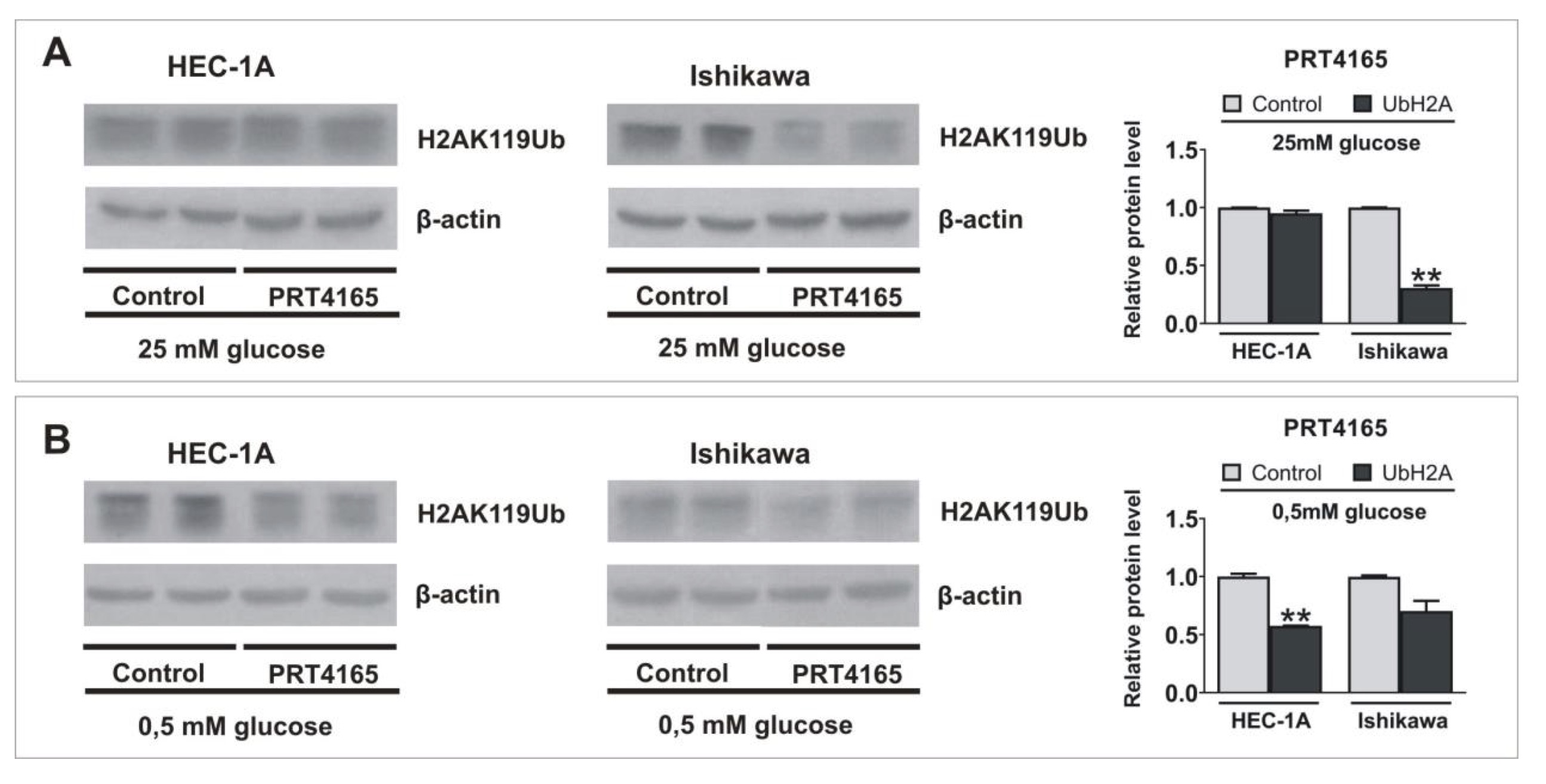
Fig. 4: Impact of inhibition of RING1/RNF2 activity on the ubiquitinylation of histone H2A lysine 119. HEC-1A and Ishikawa cells were treated with IC50 concentration of PRT4165 in standard conditions (A) and hypoglycemia (B) for 48 h. H2AK119Ub level was analyzed by Western blot. The intensity of bands corresponding to proteins after Western blot was analyzed by densitometry. The results are shown as the fold change of proteins levels of treated cells vs. control (untreated cells) in HEC-1A and Ishikawa cells.
Impact of PRT4165 and autophagy inhibitors HCQ and Lys05 on the viability of cells
The viability of cells treated with PRT4165 alone or in combination with autophagy inhibitors was estimated using the MTT test (Fig. 5). First, the IC50 and IC25 were estimated for each compound and then cells were treated with one concentration of one compound and different concentrations of the other compound. To determine the interaction between compounds the combination index (CI) was calculated. The IC50 and IC25 values are shown in Table 1. The results showed that Ishikawa cells were much more sensitive to PRT4165 and autophagy inhibitors as evidenced by the much lower concentration of compounds necessary to obtain a 50% decrease in viability. Moreover, the Lys05 inhibitor was more effective than HCQ. The calculated combination index for each treatment is shown in Table 2. The results suggest the antagonistic effect of compounds.
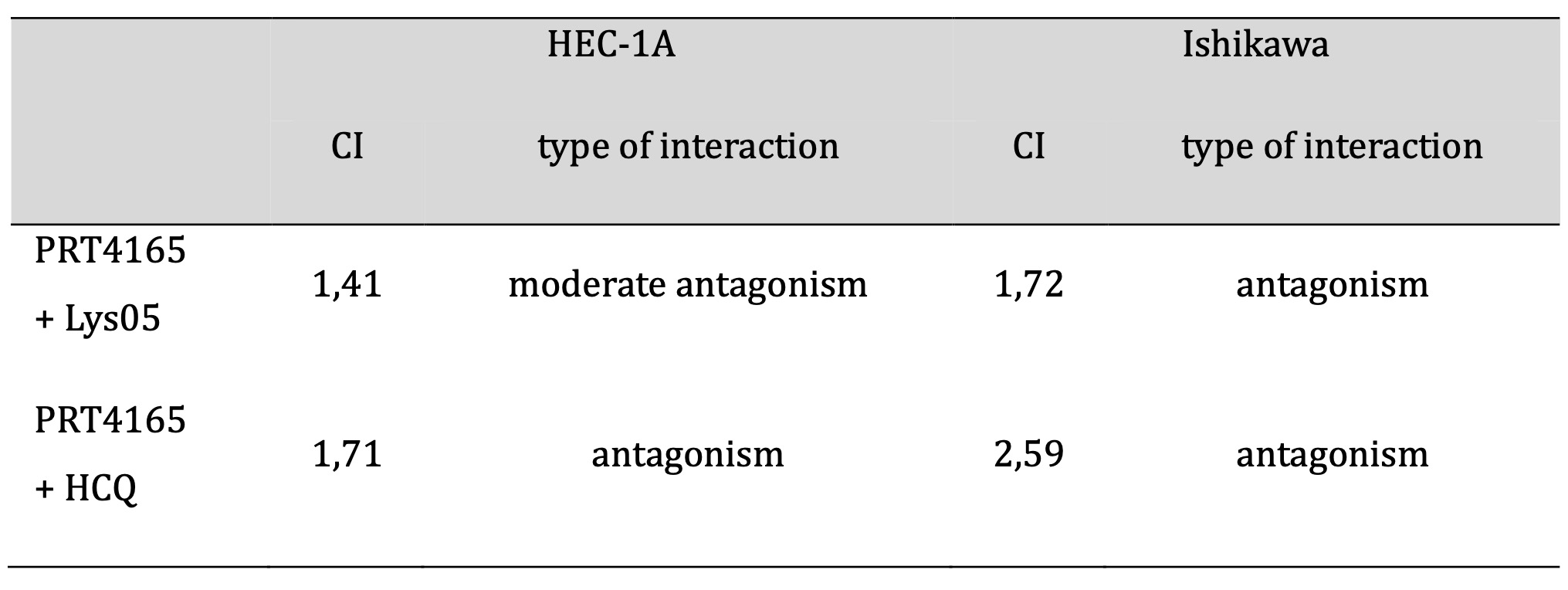
Table 2: Interactions between PRT4165 and autophagy inhibitors in HEC-1A and Ishikawa cell lines
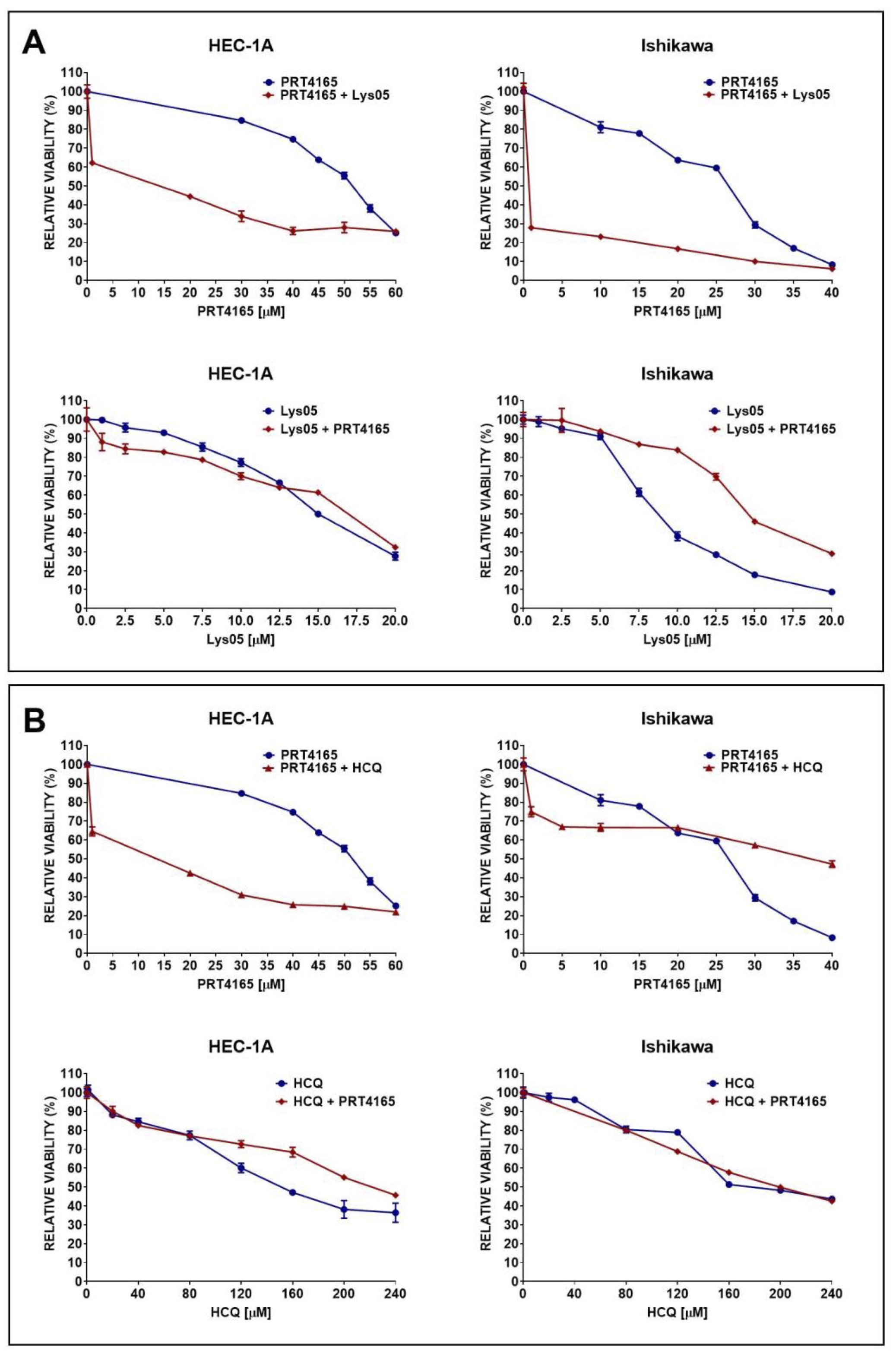
Fig. 5: Impact of PRT4165 and autophagy inhibitors Lys05 (A) or HCQ (B) and combination of compounds on the viability of HEC-1A and Ishikawa cells after 48 h treatment. The Fig. shows the means ± SEM for three experiments performed in triplicate; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
Effect of PRT4165 and Lys05 on apoptosis
The effect of PRT4165 and Lys05 on apoptosis was analyzed by flow cytometry using fluorescent dyes (Fig. 6). Cells treated with camptothecin were used as positive control and untreated or treated with DMSO cells were negative control. The results showed that PRT4165 and Lys05 used alone caused an increase in the percentage of cells in early and late apoptosis compared to control HEC-1A cells. However, the percentage of cells treated with a combination of compounds in apoptosis was lower compared to a positive control (camptothecin) and PRT4165 treatment. In the case of Ishikawa cells the effect of compounds was not pronounced.
Since the effect of PRT4165 on apoptosis was not profundal in Ishikawa cells the expression of proteins involved in necroptosis was checked. Fig. 7 shows the results of the immunodetection of RIP and the phosphorylated form of RIP after treatment of cells with PRT4165, Lys05, or a combination of both compounds. The results show that PRT4165 increases of amount proteins involved in the necroptosis process, i.e. RIP and MLKL. A similar effect, but in less extent is observed after treatment with Lys05. However, after the treatment of cells with the combination of compounds the effect is reduced. An increased ratio of pRIP/RIP is seen only in Ishikawa cells.
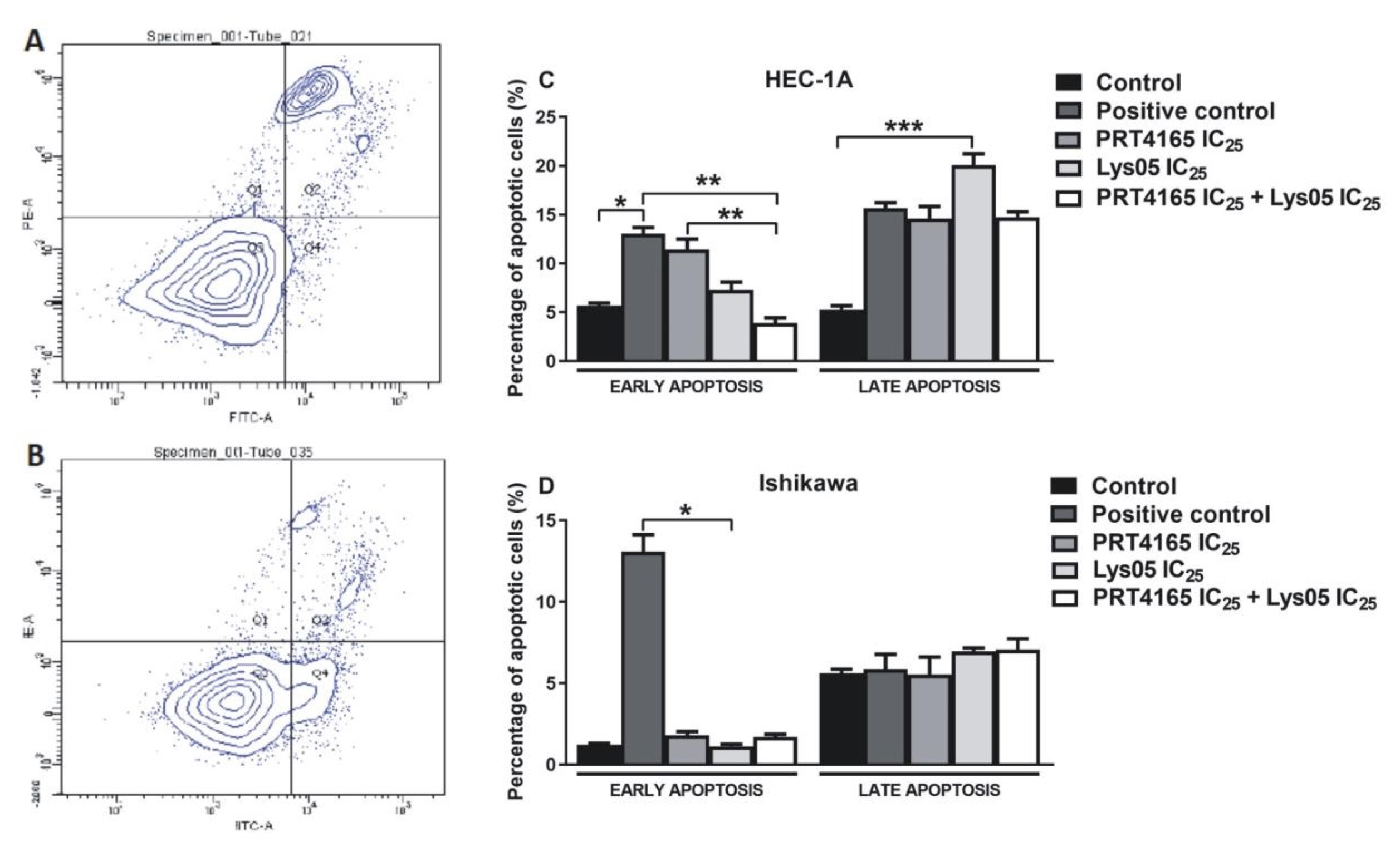
Fig. 6: Impact of PRT4165, Lys05 and its combination on apoptotic cell level. Sample histograms showing the content of live cells (Q3), necrotic cells (Q1), and in the early (Q4) and late (Q2) apoptosis after treatment with compounds for HEC-1A (A) and Ishikawa (B) cells. Percentage of cells in early and late apoptosis after 48 h treatment with PRT4165 and Lys05 and its combination. The Fig. shows the means ± SEM for three experiments performed in triplicate; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
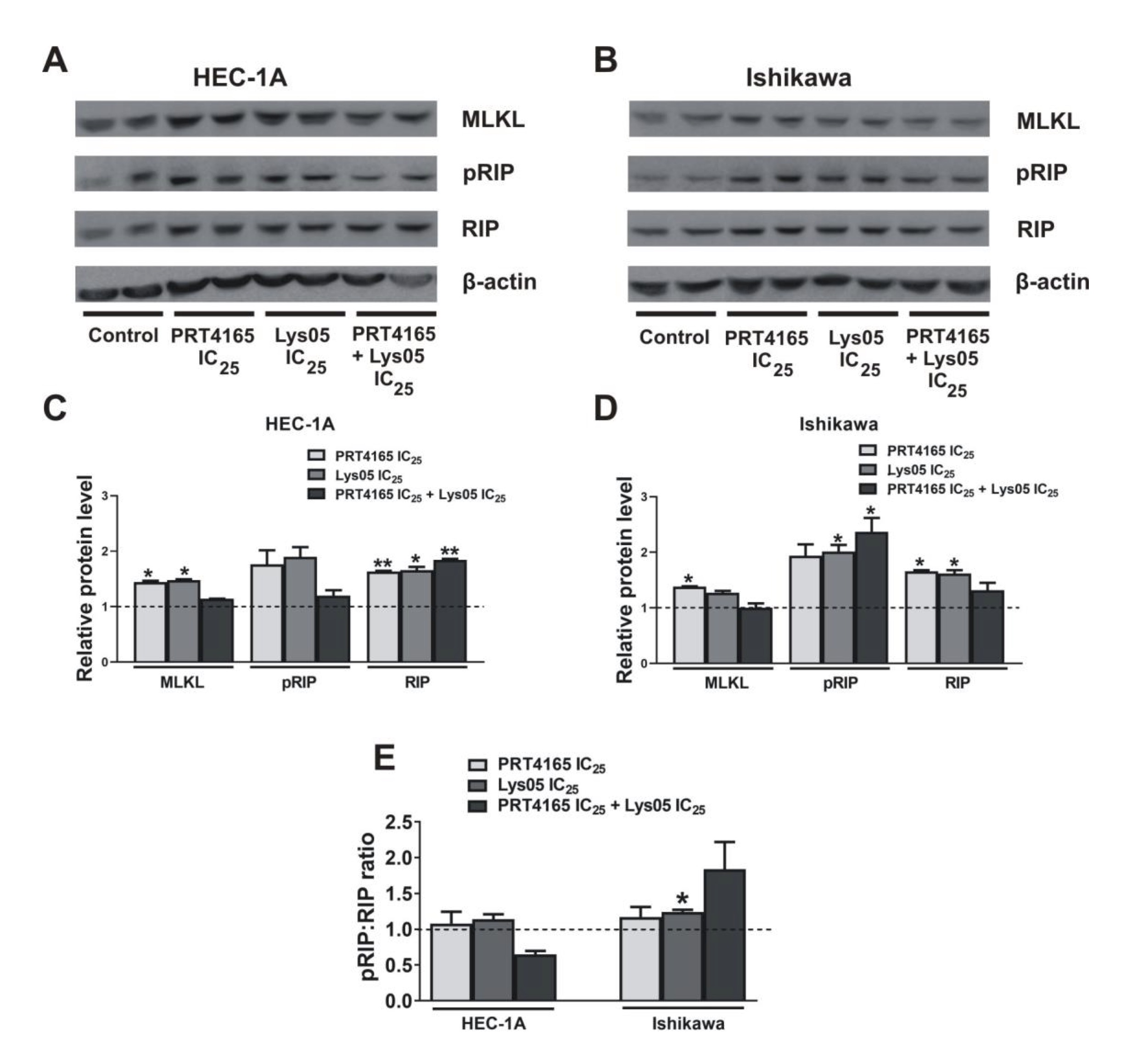
Fig. 7: Expression of proteins involved in necroptosis after treatment with PRT4165 and Lys05 in HEC-1A and Ishikawa cells. Representative immunodetection analysis of phospho-RIP, RIP and MLKL 48 h after treatment with IC25 concentration of PRT4165 (40 µM for HEC-1A, 16 µM for Ishikawa), IC25 concentration of Lys05 (10,5 µM for HEC-1A, 6,4 µM for Ishikawa) and combination of PRT4165 and Lys05 in IC25 concentration. The Fig. shows the results for two replicates. The intensity of bands corresponding to proteins after Western blot was analyzed by densitometry. The results are shown as the fold change of proteins levels of treated cells vs. control (untreated cells) in HEC-1A (C) and Ishikawa (D) cells. The phospho:RIP ratio was defined (E). The Fig. shows the means ± SEM for three experiments performed in triplicate; *p values of < 0.05; **p values of 0.01; ***p values of 0.001.
Discussion
There is not much data concerning the role of RINGs in the autophagy process. RING1A/B activity has been linked with autophagy only via AMBRA1 protein [16]. The results of the present study indicate that PRT4165 affects the expression of several genes involved in the autophagy process in endometrial cancer cells. PRT4165 is a small compound that inhibits the two E3 ligase paralogues, RING1A and RING1B and it can be used to inhibit H2A ubiquitination in cells [18]. In the presence of PRT4165 the ubiquitination of H2A histone may be completely inhibited despite RING1 or RNF2 being responsible for modification. Our results showed that in Ishikawa cells PRT4165 caused an increased amount of LC3B form and a decreased amount of p62, which are recognized as autophagy markers. During cell autophagy, the free LC3-I in the cytoplasm is converted to the lipidated active form, LC3-II (LC3B) and the level of this marker can reflect the degree of autophagy in cells. The ubiquitin- and LC3-binding protein p62 regulates the formation of protein aggregates and is removed by autophagy. The intracellular level of p62 depends on transcriptional regulation and post-transcriptional degradation in the autophagy process. During inhibition of autophagy, there is an accumulation of p62, and autophagy induction causes decreased level of p62 [19]. Thus, PRT4165 induces autophagy in Ishikawa cells but not in HEC1A cells. In mammalian cells, the main regulators of autophagy are hormones and nutritional factors. The impact of amino acids and insulin and their signaling pathways are well known but the effect of glucose deficiency as an autophagy inducer is less known. The results of these studies showed that in low glucose the expression of LC3A/B and p62 is increased both at mRNA and protein levels. Interestingly, in hypoglycemia, there is a lower amount of RING1A/RING1B proteins in cells, but the amount of mRNA level is higher, which may suggest increased degradation of protein or translation inhibition. In Ishikawa cells lower level of p62 is accompanied by an increased mRNA level, but there is not increase of LC3B, which is seen in cells growing in high glucose. Thus, PRT4165 in contrast to hyperglycemia has no effect on autophagy in hypoglycemia. In HEC-1A growing in hypoglycemia, treatment with PRT4165 does not increase the level of LC3B. This may suggest that PRT4165 depletion is less effective in low-glucose in autophagy induction. It may be associated with the fact that there is a general tendency of cancer cells to be less sensitive to therapeutic compounds for example cytostatic compounds [20, 21]
It is suggested that alterations of autophagy may play an important role in cancer, although the role of autophagy in the carcinogenesis process is ambiguous. It is believed that on the one hand, it prevents neoplastic transformation, but on the other, it promotes tumor growth. The extent autophagy may cause cell death. Autophagosomal structures may be used as specific scaffolding for apoptosis or necroptosis induction [22]. Thus, induction of early steps of autophagy with simultaneous inhibition of lysosomes may be a potential inducer of both processes. Although autophagy may be capable of ultimate cell killing when allowed to reach its limit, it is also thought to be a temporary survival mechanism under stress conditions, and inhibiting autophagy can either promote or inhibit cell death depending on the conditions and agents used [22].
A broad range of developed autophagy inhibitors have shown good preclinical results with no or low toxicity issues and with powerful anticancer activity. Clinical translations however so far been unsuccessful due to the rapid clearance, poor accumulation in the tumor sites, and side effects associated with high doses [23]. A broadly analyzed lysosome inhibitor HCQ reached phase 4 clinical trials but never obtained market approval for cancer indication. However, its extensive study helped to design and development of new potent autophagy inhibitors [23]. Targeting the autophagy process to fight cancer is still a promising strategy, especially in the framework of combination. Fukuda et al. showed that treatment of Ishikawa cells with resveratrol reduces viability and increases apoptosis of cells and usage of HCQ, potentates this effect [24]. Similarly, usage of HCQ increased the induction of apoptosis and autophagy caused by chrysin in HEC-1A and Ishikawa cells [25]. A combination of HCQ and tamoxifen against breast cancer cells with expression of estrogen receptors is more effective than each compound separately [26].
In this study, we used PRT4165 alone or in combination with lysosomes inhibitors HCQ and Lys05. Lys05 is a bivalent form of HCQ, having more potent antitumor activity both in vitro and in vivo than HCQ as a single agent in multiple human cancer cell lines and xenograft models [27]. It has been shown that the usage of anticancer drugs in combination with Lys also potentiates their efficacy [28]. The results of our study showed that PRT4165 reduces the viability of endometrial cancer cells, and Ishikawa cells are more sensitive to inhibitors. However, the combination of PRT with Lys05 or HCQ gives a mostly antagonistic effect. Thus, such a combination does not seem to be a good anticancer strategy. Evaluation of the effect of PRT4165 on apoptosis by flow cytometry showed an increase in the percentage of apoptotic cells mostly in the HEC-1A cells. However, the usage of PRT4165 inhibitor with the concomitant use of autophagy inhibitor (Lys05) has been shown to reduce the efficacy of PRT4165. Interestingly, the effect of PRT4165 is different in both cell types. The model of possible differences in PRT4165 action in HEC-1A and Ishikawa cells is shown in Fig. 8. The two endometrial carcinoma cell lines Ishikawa and HEC-1A differ with respect to the constitutive activity of the PI3-K pathway: Ishikawa cells are phospho-Akt-positive due to mutated PTEN status, whereas HEC-1A cells harbor a wild-type PTEN protein [29]. Akt is well known for its antiapoptotic activity when overexpressed or overactivated [30]. However, inhibiting components of the PI3K–Akt pathway often do not induce substantial apoptosis without additional proapoptotic insults. Thus, autophagy may be an important alternative mechanism of cell death.
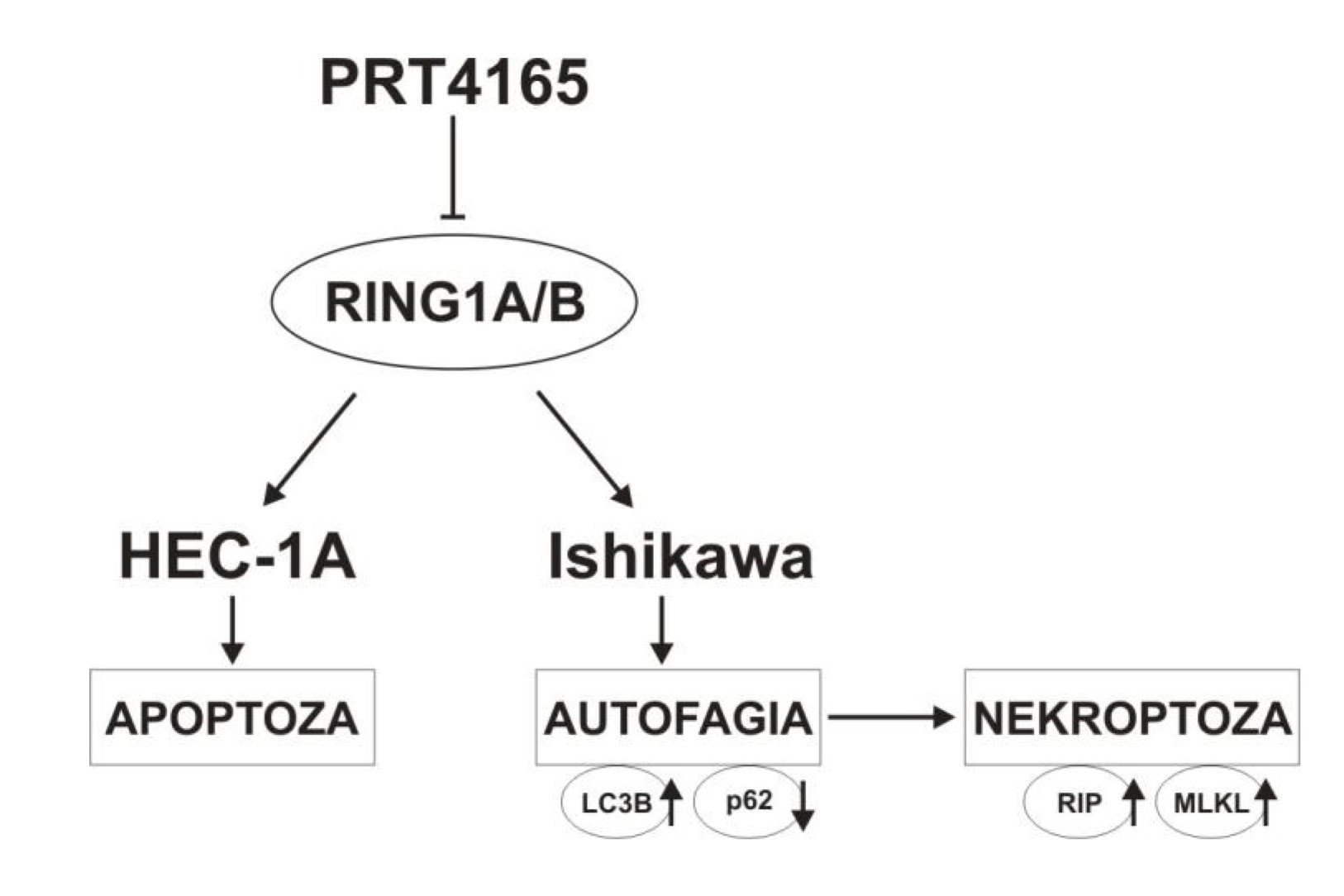
Fig. 8: The model of PRT4165 action in HEC-1A and Ishikawa cells.
Conclusion
The results of this study indicate the importance of the RINGs in the process of autophagy and apoptosis in endometrial cancer cells. PRT4165 inhibitor causes changes in the expression of ATG genes and has an anticancer efficacy by inducing autophagy or apoptosis but its influence depends on the type of cells. However, the anticancer activity of PRT4165 was lower when it was used in combination with autophagy inhibitors, suggesting that such a combination is not a promising anticancer strategy and further potentially more effective combinations of compounds should be sought.
Acknowledgements
Author Contributions
A.S. conceptualization, performing experiments, funding acquisition, writing—original draft preparation; K.K. performing experiments; A.K. conceptualization, super-vision, writing—review and editing; All authors have read and agreed to the published version of the manuscript.
Funding sources
This research was funded by a grant from the National Science Centre of Poland UMO-2018/31/N/NZ5/01046.
Statement of Ethics
The authors have no ethical conflicts to disclose
Disclosure Statement
The authors have no conflicts of interest to declare
References
| 1 | Moore K, Brewer MA: Endometrial Cancer: Is This a New Disease? Am Soc Clin Oncol Educ Book 2017;37:435-442.
https://doi.org/10.1200/EDBK_175666 |
| 2 | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 2018;68: 394-424.
https://doi.org/10.3322/caac.21492 |
| 3 | Nuñez-Olvera SI, Gallardo-Rincón D, Puente-Rivera J, Salinas-Vera YM, Marchat LA, Morales-Villegas R, López-Camarillo C: Autophagy Machinery as a Promising Therapeutic Target in Endometrial Cancer. Front Oncol 2019, 9,1326.
https://doi.org/10.3389/fonc.2019.01326 |
| 4 | Das CK, Mandal M, Kögel D: Pro-survival autophagy and cancer cell resistance to therapy. Cancer Metastasis Rev 2018;37:749-766.
https://doi.org/10.1007/s10555-018-9727-z |
| 5 | Rakesh R, PriyaDharshini LC, Sakthivel KM, Rasmi RR: Role and regulation of autophagy in cancer. Biochim Biophys Acta. Mol Basis Dis 2022;1868:166400.
https://doi.org/10.1016/j.bbadis.2022.166400 |
| 6 | Fukuda T, Wada-Hiraike O: The Two-Faced Role of Autophagy in Endometrial Cancer. Front Cell Dev Biol 2022;10:839416.
https://doi.org/10.3389/fcell.2022.839416 |
| 7 | Galluzzi L, Pietrocola F, Levine B, Kroemer G: Metabolic control of autophagy. Cell 2014;159:1263-1276.
https://doi.org/10.1016/j.cell.2014.11.006 |
| 8 | Ariosa AR, Lahiri V, Lei Y, Yang Y, Yin Z, Zhang Z, Klionsky DJ: A perspective on the role of autophagy in cancer. Biochim Biophys Acta. Mol. Basis Dis 2021;1867:166262.
https://doi.org/10.1016/j.bbadis.2021.166262 |
| 9 | Das S, Shukla N, Singh SS, Kushwaha S, Shrivastava R: Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis 2021;26:512-533.
https://doi.org/10.1007/s10495-021-01687-9 |
| 10 | Tanida I, Ueno T, Kominami E: LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004;36:2503-2518.
https://doi.org/10.1016/j.biocel.2004.05.009 |
| 11 | Shu F, Xiao H, Li Q, Ren XS, Liu, ZG, Hu BW, Wang HS, Wang H, Jiang GM: Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct Target Ther 2023;8:32.
https://doi.org/10.1038/s41392-022-01300-8 |
| 12 | Blackledge NP, Klose RJ. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol 2021;22:815-833.
https://doi.org/10.1038/s41580-021-00398-y |
| 13 | Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y: Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004;431:873-878.
https://doi.org/10.1038/nature02985 |
| 14 | Xu J, Li L, Shi P, Cui H, Yang L: The Crucial Roles of Bmi-1 in Cancer: Implications in Pathogenesis, Metastasis, Drug Resistance, and Targeted Therapies. Int J Mol Sci 2022;23:8231.
https://doi.org/10.3390/ijms23158231 |
| 15 | Fimia GM, Di Bartolomeo S, Piacentini M, Cecconi F: Unleashing the Ambra1-Beclin 1 complex from dynein chains: Ulk1 sets Ambra1 free to induce autophagy. Autophagy 2011;7:115-117.
https://doi.org/10.4161/auto.7.1.14071 |
| 16 | Xia P, Wang S, Huang G, Du Y, Zhu P, Li M, Fan Z: RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res 2014;24:943-958.
https://doi.org/10.1038/cr.2014.85 |
| 17 | Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70:440-446.
https://doi.org/10.1158/0008-5472.CAN-09-1947 |
| 18 | Ismail IH, McDonald D, Strickfaden H, Xu Z, Hendzel MJ : A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J Biol Chem 2013;288:26944-26954.
https://doi.org/10.1074/jbc.M113.461699 |
| 19 | Hönscheid P, Datta K, Muders MH: Autophagy: detection, regulation and its role in cancer and therapy response. Int J Radiat Biol 2014;90:628-635.
https://doi.org/10.3109/09553002.2014.907932 |
| 20 | Bhattacharya B, Low SH, Soh C, Kamal Mustapa N, Beloueche-Babari M, Koh KX, Loh J, Soong R: Increased drug resistance is associated with reduced glucose levels and an enhanced glycolysis phenotype. Br J Pharmacol 2014;171:3255-3267.
https://doi.org/10.1111/bph.12668 |
| 21 | Park GB, Jeong JY, Choi S, Yoon YS, Kim D: Glucose deprivation enhances resistance to paclitaxel via ELAVL2/4-mediated modification of glycolysis in ovarian cancer cells. Anticancer Drugs 2022;33:e370-e380.
https://doi.org/10.1097/CAD.0000000000001215 |
| 22 | Khandia R, Dadar M, Munjal A, Dhama K, Karthik K, Tiwari R, Yatoo MI, Iqbal HMN, Singh KP, Joshi SK: A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019;8:674.
https://doi.org/10.3390/cells8070674 |
| 23 | Bestion E, Raymond E, Mezouar S, Halfon P: Update on Autophagy Inhibitors in Cancer: Opening up to a Therapeutic Combination with Immune Checkpoint Inhibitors. Cells 2023;12:1702.
https://doi.org/10.3390/cells12131702 |
| 24 | Fukuda T, Oda K, Wada-Hiraike O, Sone K, Inaba K, Ikeda Y, Makii C, Miyasaka A, Kashiyama T, Tanikawa M: Autophagy inhibition augments resveratrol-induced apoptosis in Ishikawa endometrial cancer cells. Oncol Lett 2016;12:2560-2566.
https://doi.org/10.3892/ol.2016.4978 |
| 25 | He Y, Shi Y, Yang Y, Huang H, Feng Y, Wang Y, Zhan L, Wei B: Chrysin induces autophagy through the inactivation of the ROS mediated Akt/mTOR signaling pathway in endometrial cancer. Int J Mol Med 2021;48:172.
https://doi.org/10.3892/ijmm.2021.5005 |
| 26 | Cook KL, Wärri A, Soto-Pantoja DR, Clarke PA, Cruz MI, Zwart A, Clarke R: Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin Cancer Res 2014;20:3222-3232.
https://doi.org/10.1158/1078-0432.CCR-13-3227 |
| 27 | Amaravadi RK, Winkler J D: Lys05: a new lysosomal autophagy inhibitor. Autophagy 2012;8:1383-1384.
https://doi.org/10.4161/auto.20958 |
| 28 | Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, Raghavendra AS, Zhao Y, Bashour SI, Ibrahim NK: CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun 2017;8:15916.
https://doi.org/10.1038/ncomms15916 |
| 29 | Wan X, Li J, Xie X, Lu W: PTEN augments doxorubicin-induced apoptosis in PTEN-null Ishikawa cells. Int J Gynecol Cancer 2007;17:808-812.
https://doi.org/10.1111/j.1525-1438.2007.00890.x |
| 30 | Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B: PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer 2023;22:138.
https://doi.org/10.1186/s12943-023-01827-6 |