Original Article - DOI:10.33594/000000663
Accepted 31 August 2023 - Published online 27 September 2023
Activity of Lysosomal ABCC3, ABCC5 and ABCC10 is Responsible for Lysosomal Sequestration of Doxorubicin and Paclitaxel-Oregongreen488 in Paclitaxel-Resistant Cancer Cell Lines
bBio-Med-Chem Doctoral School of the University of Lodz and Lodz Institutes of the Polish Academy of Sciences, University of Lodz, Banacha 12/16, 90-237 Lodz, Poland,
cLaboratory of Microscopic Imaging and Specialized Biological Techniques, Faculty of Biology and Environmental Protection, University of Lodz, Banacha 12/16, 90-237 Lodz, Poland.
Keywords
Abstract
Background/Aims:
Cancer cell multidrug resistance induced by paclitaxel contributes to the high failure rates of chemotherapy and relapse of the disease. Several mechanisms have been described that underlie the observed resistance, including the overexpression of ABCB1 (P-glycoprotein), which represents an ATP-binding cassette (ABC) transmembrane protein, and its functional occurrence in lysosomal membranes is linked to drug accumulation in these organelles.Methods:
Using clinically-relevant models of paclitaxel-resistant triple-negative breast cancer and non-small cell lung cancer cell lines, we provide evidence for the role of ABCC subfamily members in the lysosomal sequestration of drugs in multidrug resistant phenotypes. Proteins expression level and its cellular localisation was measured using Western Blot and confocal microscopy. Drug accumulation was analysed by confocal microscopy and flow cytometry. Drug cytotoxicity was tested using resasurin assay and anexin V propidium iodide staining.Results:
Regardless of the alteration in gene expression, paclitaxel induced the intracellular redistribution of ABCC3, ABCC5 and ABCC10 and their enrichment in lysosomes. The use of ABCC inhibitors and transient silencing of these three genes limited the accumulation of doxorubicin and paclitaxel-OregonGreen488 in lysosomes, while having little impact on the total drug level inside cells. The cancer cells were also sensitized to various structurally unrelated chemotherapeutics of differing acidity.Conclusion:
The results suggest that lysosome membranes anchored ABCC proteins which remained functionally active and were capable to load chemotherapeutics into lysosomes in paclitaxel-resistant cancer cells. Therefore, targeting of lysosomal ABCC transporters may help to overcome paclitaxel-induced resistance by reducing the accumulation of drugs in lysosomes.Introduction
Paclitaxel (PTX) is an important first-line drug for the treatment of certain breast and advanced non-small cell lung cancers (NSCLC) due to the lack of specific markers for targeted or personalized medicine. Over 85% of lung cancer patients are diagnosed with non-small cell lung cancer (NSCLC), 60% of whom a standard chemotherapy regime is administered due to the advanced stage of disease [1–3]. Similarly, adjuvant and neoadjuvant chemo drugs are given particularly for metastatic breast cancer and triple-negative breast cancer patients (TNBC), where the cells lack estrogen, progesterone and HER2 receptors [4]. Paclitaxel is a tubulin-binding compound promoting accumulation of tubulin dimers and stabilizing microtubule fibers, which prevent proper spindle formation, arresting the cell cycle in mitosis and inducing apoptosis [5]. However, paclitaxel-induced resistance leads to treatment failure and tumor recurrence. The most frequently referred mechanisms of resistance include altered apoptotic pathways, increased drug metabolism, mutations and changes in tubulin composition, reduced drug uptake and overexpression of the pumps that actively transport the drug out of the cells [5, 6]. The latter group is represented by the ATP-binding cassette (ABC) proteins, using energy from ATP hydrolysis to translocate substances across the cell membrane. They are characterized by relatively low substrate specificity and remove a wide range of structurally unrelated compounds from cells, contributing to increased multidrug resistance (MDR) [7]. These transporters are classified into seven subfamilies (ABCA–ABCG) by the Human Genome Organization. MDR in paclitaxel-resistant human breast, lung and ovary cancers was associated with the overexpression of ABCB1 (P-glycoprotein), whereas ABCG2 (BCRP, Breast Cancer Resistance Protein) was reported in the last two tumor types [8]. Clinical trials that combined paclitaxel with P-glycoprotein inhibitors such as CBT-1 to treat solid tumors and with PSC 833 in breast, renal, lymphoma and ovarian cancer therapy were completed but not continued [9, 10]. Other ABCC subfamily members, such as ABCC1/MRP1, ABCC2/MRP2, ABCC3/MRP3, ABCB4/MDR3 were found up-regulated to various extents in paclitaxel-resistant breast cancer sublines [11], whereas the FOXM1-ABCC5 axis was shown to contribute to paclitaxel resistance in nasopharyngeal carcinoma cells [12]. Inhibitors of ABCC1 (PAK-104P), ABCB1 (LY329146) and Biricodar, which inhibits ABCC1, ABCB1 and other transporters of ABC group, have been shown to reverse paclitaxel resistance [13].
In the case of weakly basic drugs, their accumulation in sequestering organelles can be explained by changes in the intracellular pH gradient. The pH of the lumen of lysosomes is significantly lower in MDR cells, resulting in an increased pH difference between the lysosome and the cytosol. This may be a driving force for the accumulation of slightly basic anticancer agents such as doxorubicin, daunorubicin or mitoxantron (pKa = 8.3) into acidic organelles [14]. In response to repeated exposure to paclitaxel, lung cancer RERF-LC-KJ accumulated more of Oregon Green® 488 conjugated paclitaxel in the lysosomal and extra-lysosomal compartments of cytoplasm than in other cell lines [15]. This study suggested that the changes in the subcellular localization of drugs may contribute to the development of multidrug resistance. A new line of research confirms the role of lysosomes in subcellular drug sequestration and hence, limitation of the anticancer agents in reaching intracellular targets [16, 17]. It has been demonstrated that certain ABC transporters, including ABCA2-3, ABCB1, ABCC1-2, or ABCG2, sequester anti-cancer drugs into (early/late) endo- or lysosomes [16]. In the multidrug resistant human leukemic cell line HL-60 the interplay between passive daunorubicin influx into lysosomes, as well as active ABCC1-dependent sequestration of a zwitterionic molecule, sulforhodamine 101, in the Golgi apparatus was reported [18]. Furthermore, lysosomotropic weak bases such as ammonium chloride or chloroquine, as well as P-glycoprotein inhibitors (valspodar or elacridar) and silencing, markedly increased the toxicity of P-glycoprotein substrates in cervical and ovary cancers, that were characterized by overexpression of LAMP2-colocalized ABC transporters [19]. In addition to the plasma membrane, ABCC1 was seen in endocytic vesicles, perinuclearly located lysosomes and trans-Golgi vesicles [20], where its activity on intracellular vesicles was sufficient to confer a drug resistance phenotype of HeLa cells, however ABCG2 was also found in lysosomal membranes of these cells [21]. Other ABCC proteins such as ABCC2, ABCC3 and ABCC4 were also detected in crude lysosomes of some non-resistant cell lines [22], but their role in lysosome-based multidrug resistance has not been supported by experimental evidence. Although recent proteomic datasets have classified some ABC proteins as lysosomal proteins, some controversy remains as to whether their endolysosomal localization is regular or only intermediate along the trafficking pathway (with synthesis in ER/Golgi, trafficking/recycling in endosomes and degradation in lysosomes) [23].
In this study we aimed to identify the ABCC subfamily members present in the lysosomal membrane and test their possible contribution to the sequestration of chemotherapy drugs in organelles. Use was made of clinically-relevant models of paclitaxel resistant phenotypes of non-small lung cancer (A549) and triple-negative breast cancer (MDA-MB-231), searching for subcellular localization of particular ABCC proteins. To give a readout of their activity in the lysosomal membrane we traced their impact on paclitaxel accumulation conjugated with OregonGreen488, which exists as a highly polar dianion at physiological pH. The probe increases paclitaxel polarity by almost 1000-fold, thereby substantially limiting its passive diffusion across membranes and making the lysosome loading entirely dependent on membrane transporters such as ABC proteins [24]. We also used the weak base doxorubicin, which is a known substrate of ABC membrane transporters and whose passive diffusion into lysosomes raises concerns about ABCB1-dependent lysosomal sequestration [21].
Materials and Methods
Materials
Non-small-cell lung cancer cell line A549 was purchased from ATCC. Breast cancer cell line MDA-MB-231 was purchased from Sigma Aldrich. DMEM High Glucose w/ L-Glutamine w/ Sodium Pyruvate, fetal bovine serum and antibiotics (penicillin and streptomycin) were from Biowest (CytoGen, Zgierz, Poland). Resazurin sodium salt, doxorubicin hydrochloride, cisplatin, paclitaxel, etoposide, ammonium chloride (A4514), MK-571 (M7571), Lysosome Isolation Kit (LYSISO1), Anti-LAMP1 antibody (SAB3500285) were from Sigma Aldrich (Poznan, Poland). Nunc™ Lab-Tek™ Chamber Slide were ordered in Biokom (Janki/Warsaw, Poland). Anti-ABCC5 antibody (sc-376965) and siRNA Control (sc-37007) were purchased from Santa Cruz Biotechnology (AMX, Lodz, Poland). Silencer Select Pre-designated siRNA: siABCC3 (#16602), siABCC5 (#19553) and siABCC10 (#40144), Lipofectamine RNAiMAX, OptiMem, SuperSignal™ West Pico Chemiluminescent Substrate, PageRuler™ Prestained Protein Ladder (#01154870), Pierce™ Protease Inhibitor Tablets (EDTA-free; PIC), Paclitaxel Oregon Green™ 488 conjugate (Flutax-2), Lysotracker™ Deep Red, BODIPY™ TR Ceramide, MitoTracker™ Red FM, SlowFade™ Glass Soft-set Antifade Mountant (with DAPI), anti-MRP3 (ABCC3) Polyclonal Antibody (PA5101482), anti-MRP5 (ABCC5) Polyclonal Antibody (PA5102678), anti-MRP10 (ABCC10) Polyclonal Antibody (PA5101678), SOD2 Antibody (1H6) (#MA1-106) were from Thermofisher Scientific (Thermofisher Scientific, Warsaw, Poland). Anti-ABCB1 (E1Y7B) Rabbit mAb (#13342), anti-ABCC1 (D7O8N) rabbit mAb (#14685), anti-ABCC3 (D8V8J) Rabbit mAb (#39909), ABCG2 (D5V2K) XP® Rabbit mAb (#42078), anti-ACTB (E4D9Z) Mouse mAb (#58169), Histone H3 Rabbit Ab (#9715), anti-rabbit IgG, HRP-linked Antibody (#7074), Anti-mouse IgG, HRP-linked Antibody (#7076), Anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor® 488 Conjugate) (#4412), Anti-rabbit IgG Fab2 (Alexa Fluor® 594 Conjugate) (#8889), Anti-mouse IgG (H+L), F(ab’)2 fragment (PE Conjugate) (#59997) were from Cell Signaling Technologies (LabJOT, Warsaw, Poland). An Annexin V Apoptosis Detection Kit with Propidium iodide was purchased from BioLegend (BioCourse.pl, Katowice, Poland).
Cell culture and treatment with inhibitors
A549 cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin (50 U/ml and 50 µg/ml, respectively) in 5% CO2. Initially, MDA-MB-231 cells were cultured in F15 medium supplemented with 15% FBS and penicillin/streptomycin (50 U/mK and 50 µg/ml, respectively) without CO2 equilibration. After 5 passages, the cells were adapted to grow in DMEM supplemented with 10% FBS and penicillin/streptomycin (50 U/ml and 50 µg/ml, respectively) in 5% CO2.
Paclitaxel at the concentration of 0.05 µM was added to cells for 48 h every 3 weeks for a total number of 6 cycles. The treatment scheme was chosen to fit the current therapeutic approaches, and drug concentration to fit the minimal paclitaxel concentration in the blood of patients [25]. The drug was washed out after 48 h and cells were cultured in the full growth medium till another dose of paclitaxel. After 6 cycles of treatment cells were froze and kept in liquid nitrogen. After thawing, one dose of paclitaxel was added to cells and all experiments were performed up to six consecutive cell passages. Drug-resistant and non-resistant cell lines were cultured under the same condition.
Ammonium chloride (25 mM) and ABCC transporter inhibitor MK-571 (25 µM) were added to cells for 2 h prior to analysis or treatment with anticancer drugs. Depending on the tested parameters, anticancer drugs were added to cells for 24 or 48 h.
Lysosome isolation
For the isolation of lysosomes destined for cytometer analysis, 2 million cells per sample were used; for Western Blot samples, 200 million cells per sample were used. The lysosome isolation was prepared according to the Lysosome Isolation Kit manufacturers protocol. Cells were trypsinized and washed two times with PBS. The cells were then resuspended in Extraction Buffer and sonicated (Bandelin Sonopuls HD2070). After every five strokes, cells were checked under a microscope to ascertain the degree of the breakage. The sonication was stopped at 80-85% of breakage. The samples were then centrifuged at 1000 x g for 10 min. The supernatant containing cells content was transferred to the new tubes and centrifuged at 20000 x g for 20 min. The pellet containing the Crude Lysosomal Fraction (mixture of mitochondria, lysosomes, peroxisomes and endoplasmic reticulum) was suspended in Extraction Buffer. Next, the samples were diluted to a solution containing 19% Optiprep Density Gradient Medium (0.505 ml Optiprep, 0.65ml Optiprep Dilution Buffer, 0.03 ml 2, 3 M Sucrose to 0.4 ml of Crude Lysosomal Fraction). To the diluted Optiprep fraction 250mM Calcium chloride was added to a final concentration of 8 mM. Next the samples were incubated on ice for 15 min and centrifuged for 5000 x g for 15 min to allow the precipitation of mitochondria, endoplasmic reticulum and peroxisomes. Finally, the supernatant containing lysosomes was transferred into fresh tubes. Lysosome purity and lysosomal membrane integrity were tested by flow cytometry after lysosome staining with Neutral Red dye, provided in the Lysosome isolation Kit. Fluorescence was read at 606 nm, the fluorescence of the lysosomotropic dye in the hydrated and acidic interior of lysosomes [26].
Formation of cell spheroids
Nunc™Lab-Tek™chamber slides were coated with faCellitate BIOFLOAT FLEX coating solution according to the manufacturer protocol. After 30 min of air-drying of the chambers within the laminar flow hood, the cells were seeded per well at a density of 20, 000 cells. Cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin (50 U/ml and 50 µg/ml, respectively) in 5% CO2 to allow spheroids formation for 21 days.
Transient gene silencing
The cells were seeded per well at a density of 100, 000 cells on the 24-well plate, 10, 000 cells on Nunc™ Lab-Tek™ Chamber Slide or 3-week spheroids were transfected using siRNA-RNAiMAX complexes according to the previously described protocol [27]. After 6 h incubation with the complexes, DMEM supplemented with 10% FBS and antibiotics was added to the desired volume and cells were grown for another 48 h to obtain transient gene silencing.
Flow cytometry
Cells were seeded into a 6-well plate at a density of 1 million cells per well. After 48 h of cell transfection, doxorubicin and paclitaxel-Oregon Green were added to the culture for another 24 h. Then lysosomes were isolated or cells were trypsynized and washed with PBS, fixed in 0.5% formaldehyde in PBS at room temperature for 15 min and transferred to fresh PBS. The lysosomes were taken directly after isolation to analysis. The fluorescence intensity was measured by a flow cytometer LSR® II (Becton Dickinson) at ex: 470/em: 595 nm/nm for anthracyclines and ex: 496 nm/em: 524 nm for paclitaxel. The cell or lysosome population was discriminated based on FSC-A and SSC-A parameters in Flowing Software 2. Intensity of the cell fluorescence was visualized on a histogram and the shift in fluorescence distribution indicated the alteration in drug distribution.
Whole-cell protein expression evaluation
For protein expression evaluation cells were lysed in RIPA buffer (supplemented with 1 mM PMSF and PIC) and sonicated (Bandelin Sonopuls HD2070); Proteins were then separated by SDS–PAGE, transferred into a nitrocellulose membrane and stained with primary antibodies (1:5000) at 4°C overnight. After subsequent staining with HRP-conjugated secondary antibodies (1:5000 for antirabbit and 1:2500 for anti-mouse antibodies; room temperature; 2 h), the signal was developed with the SuperSignal™ West Pico Chemiluminescent Substrate and pictures were acquired using ProXima 2750 (Isogen Life Science). ACTB was used as the control.
Lysosomal protein expression evaluation
Lysosomes were suspended in 5x RIPA buffer (supplemented with 1 mM PMSF and PIC) and sonicated (Bandelin Sonopuls HD2070). Lysosome lysates were then separated by SDS–PAGE, transferred into a nitrocellulose membrane and stained with primary antibodies (1:5000) at 4°C overnight. After subsequent staining with HRP-conjugated secondary antibodies (1:10, 000 for antirabbit and 1:5000 for anti-mouse antibodies; room temperature; 2 h), the signal was analyzed using the SuperSignal™ West Pico Chemiluminescent Substrate in ChemiDoc-IT2 (UVP, Meranco, Poznan, Poland). ACTB, H3 and SOD2 was used as the control.
Confocal microscopy evaluation of ABC proteins
For the confocal imaging of ABC proteins, cells were seeded on a Nunc™ Lab-Tek™ chamber slide. 2 h before cell fixation, Lysotracker™ Deep Red was added to the final volume of 75 nM and cells were incubated 1 h at 37°C. Cells were fixed with a 1% formaldehyde solution in PBS at room temperature for 15 min, permeabilized and blocked with 1% FBS solution in PBS with 0.1% TritonX-100 at room temperature for 1 h. Primary antibodies (1:400) were added in 1% BSA solution in PBS with 0.1% TritonX-100 and incubated at 4 °C overnight. A secondary antibody (1:400) was then added in 1% BSA solution in PBS with 0.1% TritonX-100 at room temperature for 2 h. After washing, the slides were mounted with SlowFade™ glass soft-set antifade mountant (with DAPI). The TCS SP8 (Leica Microsystems, Germany) with a 63x/1.40 objective (HC PL APO CS2, Leica Microsystems, Germany) was used for sample visualization. The samples were imaged with the following wavelength values for excitation and emission: 485 and 500-550 nm for Alexa Fluor® 488, 647 and 660-670 for Lysotracker Deep Red and 405 and 430-480 nm for DAPI. The fluorescence intensity and colocalization was determined in arbitrary units (a.u.) with Leica Application Suite X (LAS X, Leica Microsystems, Germany).
Confocal microscopy imaging of lysosomal proteins
For the confocal imaging of ABC proteins, cells were seeded on a Nunc™ Lab-Tek™ chamber slide. Cells were fixed with a 1% formaldehyde solution in PBS at room temperature for 15 min, permeabilized and blocked with 1% FBS solution in PBS with 0.1% TritonX-100 at room temperature for 1 h. Anti-LAMP primary antibody (1:400) was added in 1% BSA solution in PBS with 0.1% TritonX-100 and incubated at 4 °C overnight. A secondary antibody labelled with Alexa Fluor® 488 (1:400) was then added in 1% BSA solution in PBS with 0.1% TritonX-100 at room temperature for 2 h. Next anti-ABC primary antibodies (1:400) were added in 1% BSA solution in PBS with 0.1% TritonX-100 and incubated at 4 °C overnight. A secondary antibody labelled with Alexa Fluor® 546 or R-phycoerythrin (PE) (1:400) was then added in 1% BSA solution in PBS with 0.1% TritonX-100 at room temperature for 2 h. After washing, the slides were mounted with SlowFade™ glass soft-set antifade mountant (with DAPI). The TCS SP8 (Leica Microsystems, Germany) with a 100x/1.40 objective (HC PL APO CS2, Leica Microsystems, Germany) was used for sample visualization. The samples were imaged with the following wavelength values for excitation and emission: 485 and 500-550 nm for Alexa Fluor® 488, 550 and 570-580 nm for Alexa Fluor® 546, 480 and 570-580 nm for R-phycoerythrin (PE). The scans of cells were deconvolved using 3D-Deconwolution accessible in Leica Application Suite X software (LAS X, Leica Microsystems, Germany).
Confocal microscopy analysis of drug accumulation in cells
Cells seeded on Nunc™Lab-Tek™chamber slides were treated with anticancer drugs and inhibitors. Next, Lysotracker™ Deep Red (75 nM for 2 h), BODIPY™ TR Ceramide (5 µM for 30 min), or MitoTracker™ Red FM (100 nM for 30 min), was added to the culture media. After incubation and cell washing with PBS, cells were mounted with SlowFade™ glass soft-set antifade mountant (with DAPI). The TCS SP8 (Leica Microsystems, Germany) with a 63x/1.40 and 10x 0.40 DRY objectives (HC PL APO CS2, Leica Microsystems, Germany) was used for sample visualization. The samples were imaged with the following wavelength values for excitation and emission: 485 and 500-550 nm for Alexa Fluor® 488 conjugated Paclitaxel, 470 and 580-600 nm for Doxorubicin and 405 and 430-480 nm for DAPI. The fluorescence intensity was determined in arbitrary units (a.u.) with Leica Application Suite X (LAS X, Leica Microsystems, Germany).
Confocal microscopy evaluation of drug accumulation in spheroids
3-week-old 3D cell cultures were stained with Lysotracker™ Deep Red (75 nM) for 2 h. Next, spheroids were fixed with a 4% formaldehyde solution in PBS, at room temperature for 30 min. After fixation the spheroids were washed with PBS and incubated with 1 µg/ml DAPI for 30 min at room temperature. Spheroids were analyzed immediately. The TCS SP8 (Leica Microsystems, Germany) with a 63x/1.40 and 10x 0.40 DRY objectives (HC PL APO CS2, Leica Microsystems, Germany) was used for sample visualization. The samples were imaged with the following wavelength values for excitation and emission: 485 and 500-550 nm for Alexa Fluor® 488 conjugated Paclitaxel, 470 and 580-600 for Doxorubicin and 405 and 430-480 nm for DAPI. The fluorescence intensity was determined in arbitrary units (a.u.) with Leica Application Suite X (LAS X, Leica Microsystems, Germany).
Resazurin toxicity assay
The day prior to transfection/treatment, cells were seeded at a density of 2, 500 cells per well on Nunc® MicroWell™ 384-well optical bottom plates. After transfection/incubation with drugs and inhibitors, cells were incubated with the resazurin solution (5 µM) in the growth medium at 37 °C for 3 h. The fluorescence that corresponds to the metabolic activity of living cells was measured with a fluorescence microplate reader (BioTek Synergy HTX, Biokom, Poland) at excitation 530 and emission 590 nm. The fluorescence value for control cells was assumed to be 100%.
Annexin V and propidium iodide staining
For the visualization of apoptosis/necrosis induction by the combination of drugs and inhibitors, Nunc™Lab-Tek™chamber slides were coated with faCellitate BIOFLOAT FLEX coating solution according to manufacturer protocol. Cells seeded on coated chamber slides were harvested for 3-weeks to obtain spheroid formation. Cells were treated with inhibitor/ siRNA-RNAiMAx complexes and anticancer drugs. After washing with PBS, according to the manufacturer protocols, Annexin V-FITC and Propidium iodide were added to the cells in a binding buffer [28] and incubated for 30 min at room temperature. After incubation, spheroids were washed with PBS and incubated with 1µg/ml DAPI for 30 min at room temperature. Spheroids were stored in PBS. The confocal laser scanning microscopy platform TCS SP8 (Leica Microsystems, Germany) with a 10x objective (HC PL APO CS2, Leica Microsystems, Germany) was used for the imaging of samples at the following wavelength values for excitation and emission: 485 and 500-550 nm for AnnexinV-FITC, 450 and 610-620 for Propidium iodide, 405 and 430-480 nm for DAPI, using Leica Application Suite X (LAS X, Leica Microsystems). The level of baseline fluorescence was established individually for each experiment. Fluorescence intensity was determined as arbitrary units (a.u.) with the Leica Application Suite X (LAS X, Leica Microsystems, Germany).
Statistical analysis
Data are shown as mean ± standard deviation (SD). Parametric or non-parametric tests were conducted after testing the Gaussian distribution of data with the Shapiro–Wilk test. Student’s t-test or the Mann–Whitney test was used to calculate statistically significant differences between two samples, while one-way analysis of variance (ANOVA) or the Kruskal-Wallis test followed by corresponding post hoc test was carried out to compare multiple samples. Statistics were calculated using GraphPad Prism 8.01 software. Statistically significant differences were marked with * when p < 0.05, ** when p < 0.01, *** when p < 0.001.
Results
Paclitaxel resistance is associated with enrichment of some ABCC transporters in lysosomal membranes
Knowing that paclitaxel-resistant cancer cells are characterized by overexpression of the three most frequently referred ABC proteins such as ABCB1, ABCC1 and ABCG2 and that ABCB1 is localized in some intracellular organelles [7, 8,15, 16, 29–31], we tested for expression and particularly searched for intracellular redistribution of ABCC subfamily members caused by a high cell exposure to paclitaxel. Among the tested and multidrug resistance-relevant transporters, only ABCB1 and ABCC5 increased in both studied cell lines as imaged by confocal microscopy and Western Blot analysis of whole cell lysates (Fig. 1A-B, 1F-G, S1A-B, S1D, S2A-B, S2D). An elevated level of ABCC1 was found in A549 cells resistant to paclitaxel (Fig. S1A, S1D), ABCC10 in MDA-PTX (Fig. S2A, S2D), whereas ABCC3 declined in both paclitaxel-resistant cell lines (Fig. S1A, S1D, Fig. S2A, S2D). The development of drug resistance was associated with the enrichment of ABCB1, ABCC3, ABCC5 and ABCC10 in lysosomes, stained with Lysotracker™ Deep Red, as shown in confocal images and by colocalization of these proteins with lysosomes (Fig. 1A, 1C, 1D, 1F, 1H, 1I, S1A, S1C, S2A, S2C, S3), regardless of their paclitaxel-induced alteration in cellular abundance. ABCB1, ABCC3, ABCC5 and ABCC10 were further confirmed by observing the increased level of all these four proteins in the membranes of lysosomes isolated from paclitaxel-resistant phenotypes (Fig. 1E, 1J). ABCC1, which reportedly occurred in the Golgi apparatus and an intracellular vehicle in multidrug resistant cells, remained anchored in the plasma membrane.
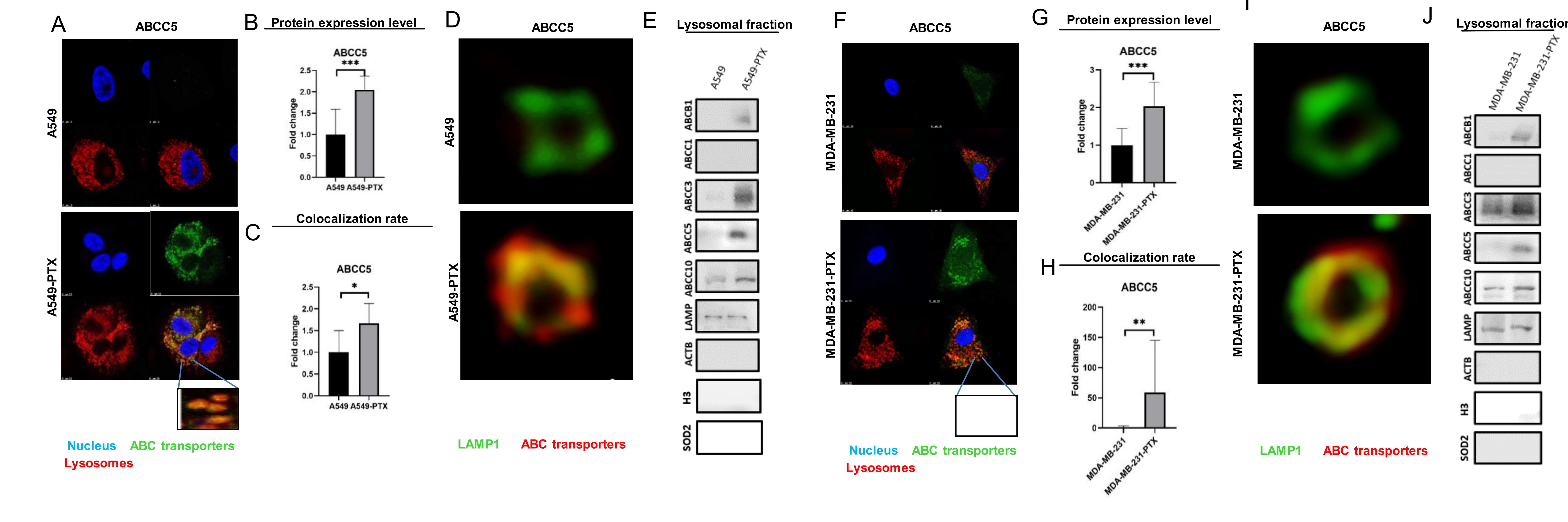
Fig. 1: Paclitaxel resistance is associated with enrichment of ABC transporters in lysosomes. A, F) Comparison of selected ABC transporter abundance and intracellular localization in non- and paclitaxel-resistant cancer cell lines A549 (A) and MDA-MB-231 (F). ABC transporters were visualized by immunocytostaining followed by confocal microscopy. Green fluorescence of ABC transporters is derived from Alexafluor488-conjugated secondary antibody, blue DNA was stained with DAPI, whereas red lysosomes with LysoTrackerTM Deep Red. (B-C, F-G) The fluorescence intensity, which corresponds to ABC protein level inside cells (B; G) and colocalization between considered proteins and lysosomes (C, H) were determined in arbitrary units (a.u.) with Leica Application Suite X. The difference between two means was tested with Student’s t-test or Mann Whitney test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001. (D,I) Confocal microscopy imaging of lysosomal proteins in non- and paclitaxel-resistant cancer cell lines A549 (D) and MDA-MB-231 (I). ABC transporters were visualized by immunocytostaining followed by confocal microscopy. Red fluorescence of ABC transporters is derived from R-phycoerythrin- labelled secondary antibody and LAMP1- green fluorescence is derived from Alexafluor488-conjugated secondary antibody. The scans of lysosomes were deconvolved using 3D-Deconwolution accessible in Leica Application Suite X software (LAS X, Leica Microsystems, Germany). (E, J) ABC transporters were detected and compared in isolated lysosomes of cancer cell lines and their paclitaxel-resistant counterparts by western blot. LAMP was used as an internal and positive control, whereas ACTB served as a control for lysosome contamination with cytoplasmic and plasma membrane fractions; SOD2 was a control for contamination with mitochondria and histone H3 served as control for lysate contamination with nuclear fraction.
Paclitaxel resistance enhances the sequestration of acidity-different compounds in lysosomes
Knowing that a highly acidic environment of lysosomes determines the passive accumulation of compounds depending on their acidity, we made use of neutral Paclitaxel Oregon Green 488 that can only enter lysosomes by active membrane transporters, and we traced its intracellular localization by confocal microscopy and co-staining of selected organelles. In parallel, intracellular distribution of doxorubicin was tested. Doxorubicin enters acidic organelles due to weak base chemical features, but it is also transported across cellular membranes by the ABC proteins, making it possible to estimate the contribution of ABC proteins in doxorubicin loading to lysosomes. As shown in Fig. 2A-D and Fig. S4A-B, gaining resistance to paclitaxel considerably changed intracellular localization of both drugs. In non-resistant cells, neutral Paclitaxel Oregon Green 488 accumulated in the cytoplasm (cytoskeleton binding), whereas doxorubicin was found in cytoplasm and to a lower extent also in the nucleus. Repeated exposure to paclitaxel enhanced the drug sequestration in lysosomes, as indicated by substantial increase in the drug-lysosome co-localization rate (Fig. 2C, 2D).
To confirm enhanced accumulation of the considered drugs in lysosomes enriched in ABCB1, ABCC3, ABCC5 and ABCC10, the fluorescence intensity in lysosomes isolated from non-resistant and paclitaxel-resistant cells exposed to Paclitaxel OregonGreen 488 and doxorubicin was compared. The median value of fluorescence distribution indicates 3- and 7-fold increase in lysosomal trapping of paclitaxel and anthracycline respectively in paclitaxel-resistant phenotypes (Fig. 2E-F).
Taking into account literature reports on the possible non-specific binding of Oregon Green 488-labeled paclitaxel to the Golgi apparatus [32] and enrichment of ABCB1 in the mitochondria membrane [29], the impact of development of paclitaxel resistance on the possible accumulation of fluorescently labeled paclitaxel in these two cellular compartments was checked (Fig. S4C-F). As shown in Fig. S4C-D, colocalization of the drug, endoplasmic reticulum and Golgi apparatus was substantially lower in the resistant phenotype of A549 cells. Based on the confocal images in Fig. S4E-F, mitochondria of resistant cells accumulated a lower amount of Paclitaxel Oregon Green 488 indicating that intracellular redistribution of ABCB1, ABCC3, ABCC5 and ABCC10 to lysosomes co-occurs with considerably enhanced Paclitaxel Oregon Green 488 and doxorubicin accumulation in these organelles.
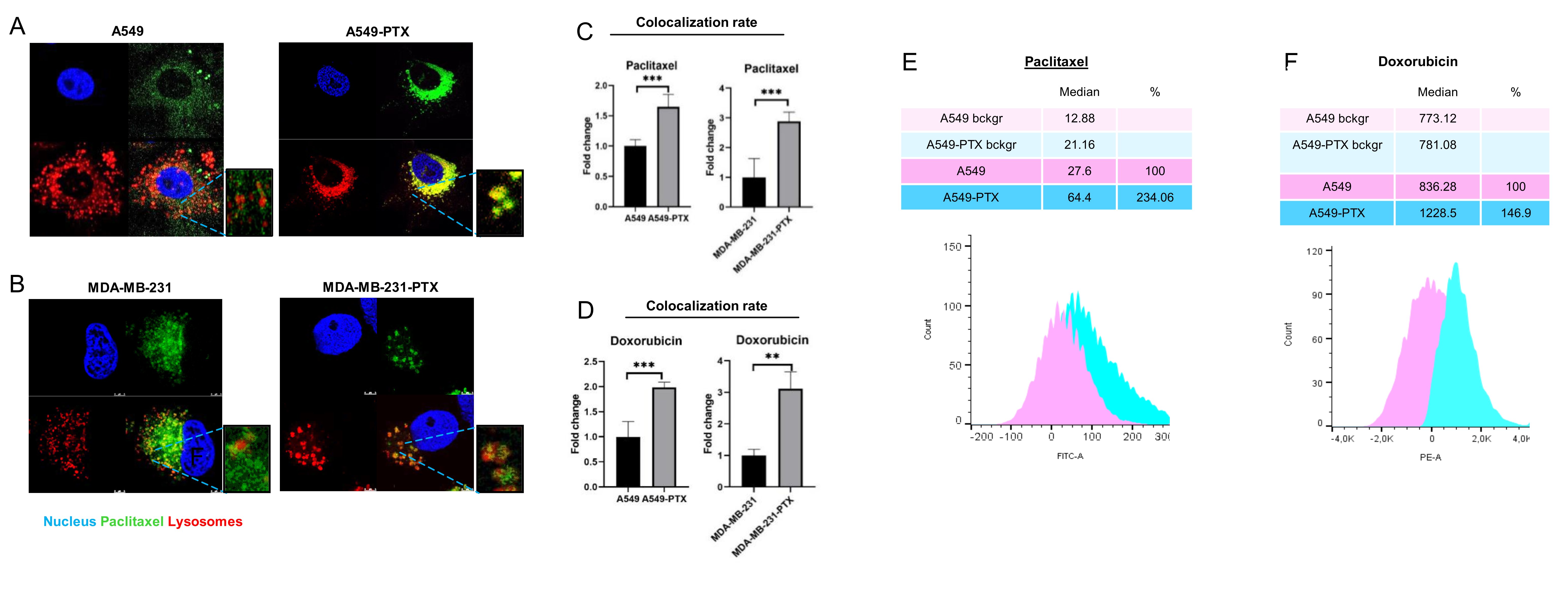
Fig. 2: Development of cancer cell resistance to paclitaxel is followed by intracellular drug redistribution to lysosomes. (A-B) Comparison of paclitaxel intracellular localization between non-resistant and paclitaxel-resistant cancer cell lines (A549 – A and MDA-MB-231 – B) after their exposure to paclitaxel for 24 h based on confocal imaging (TCS SP8, Leica Microsystems, Germany). Paclitaxel Oregon GreenTM 488 is visualized in green (0,1 µM), DNA (DAPI) in blue and lysosomes (LysoTrackerTM Deep Red) in red. (C,D) The colocalization between fluorescence of Paclitaxel Oregon GreenTM 488 (C),autofluorescence of doxorubicin (D) and LysoTrackerTM Deep Red was determined in arbitrary units (a.u.) with Leica Application Suite X. The difference between two means was tested with Student’s t-test or Mann Whitney test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001 (G-H) Comparison of doxorubicin (G) and paclitaxel (H) lysosomal uptake between non-resistant and paclitaxel-resistant A549 cancer cell line after their exposure to drugs for 24 h based on Flow Cytometry. The fluorescence intensity was measured by flow cytometer LSR® II (Becton Dickinson) at em: 595 nm/ex: 470 nm for anthracyclines and em: 524 nm/ex: 496 nm for paclitaxel. Distribution of cell/lysosomes intensity was analysed in Flowing Software 2. The analyzed cell population was discriminated based on FSC-A and SSC-A parameters. Intensity of cell fluorescence was visualized on a histogram and the shift in fluorescence distribution indicated the alteration in drug distribution.
Development of cancer cell resistance to paclitaxel is followed by ABCC transporter- dependent drug redistribution to lysosomes
Taking into account previous reports on the lysosomal function of ABCB1 [19, 33], we focused mostly on ABCC3, ABCC5 and ABCC10. Their lysosomal localization and possible impact on drug sequestration in these organelles has not been mechanistically described.
To confirm the role of lysosomal ABC transporters in lysosomal drug sequestration, the uptake of chemotherapeutics was analyzed in the presence of two inhibitors MK-571 and ammonium chloride (AC). MK-571 is a non-selective inhibitor of several ABCC-transporters, including ABCC1, ABCC2, ABCC3, ABCC5 and ABCC10 [34–40]. Due to its non-selective action against several transporters, it serves as a potential pan-ABCC inhibitor [41]. AC is a lysosomal lumen alkalizer, which increases the acidic pH of the lysosomes, thereby leading to the passage of neutral cytotoxic drugs across the lysosomal membrane and considerable release of weak bases entrapped in these organelles [33]. Ammonium chloride declined accumulation of the doxorubicin inside lysosomes in a resistant lung cancer cell line grown in a monolayer, but had weaker impact on Paclitaxel Oregon Green 488 intracellular localization (Fig. 3A-B, Fig. S5A). Interestingly, iABCC was more potent in reducing colocalization between lysosomes and both drugs, and almost completely prevented doxorubicin sequestration inside lysosomes. This indicates that the ABCC subfamily of proteins is involved in lysosomal sequestration of the studied drugs.
The role of lysosomes in drug accumulation was also confirmed in a 3D cancer cell culture treated with ammonium chloride, causing deeper penetration of Paclitaxel Oregon Green 488 and doxorubicin (Fig. 3C, Fig. S5B-D). In paclitaxel-resistant spheroids fluorescently labeled paclitaxel was entrapped in peripheral cell layers and visualized on fluorescence plots across the 3D culture. Ammonium chloride also increased the signal of both drugs in the central part of the culture, thereby allowing them to target the cells located in the inner spheroid layers. The ABCC pan-inhibitor MK-571 phenocopied the effect of ammonium chloride, thereby suggesting that ABCC subfamily of proteins entrap these two drugs in the outer layer of spheroid and likely in the lysosomes.
The functional impact of drug sequestration in the lysosomes by ABCC transporters and drug toxicity was studied by evaluating apoptotic and necrotic markers (Fig. 3D, 3E). Analysis of Annexin V intensity in 3D culture of A549-PTX spheroids confirmed a stronger apoptotic induction with the combination of drug and iABCC than with drugs alone, whereas not observing a substantial extent of necrosis. Accordingly, confocal imaging of Annexin V and propidium iodide-stained spheroids showed a similar distribution of apoptotic cells in all spheroid layers after spheroid exposure to the combination of drug and iABCC (Fig. 3F, Fig. S5E). A strong Annexin V signal in 3D cultures treated with the drug alone was observed in the outer parts of spheroid. All these results suggest that drug entrapment by lysosomal ABCC transporters prevents the drug reaching a pharmacological target and hence, reduces drug cytotoxicity.
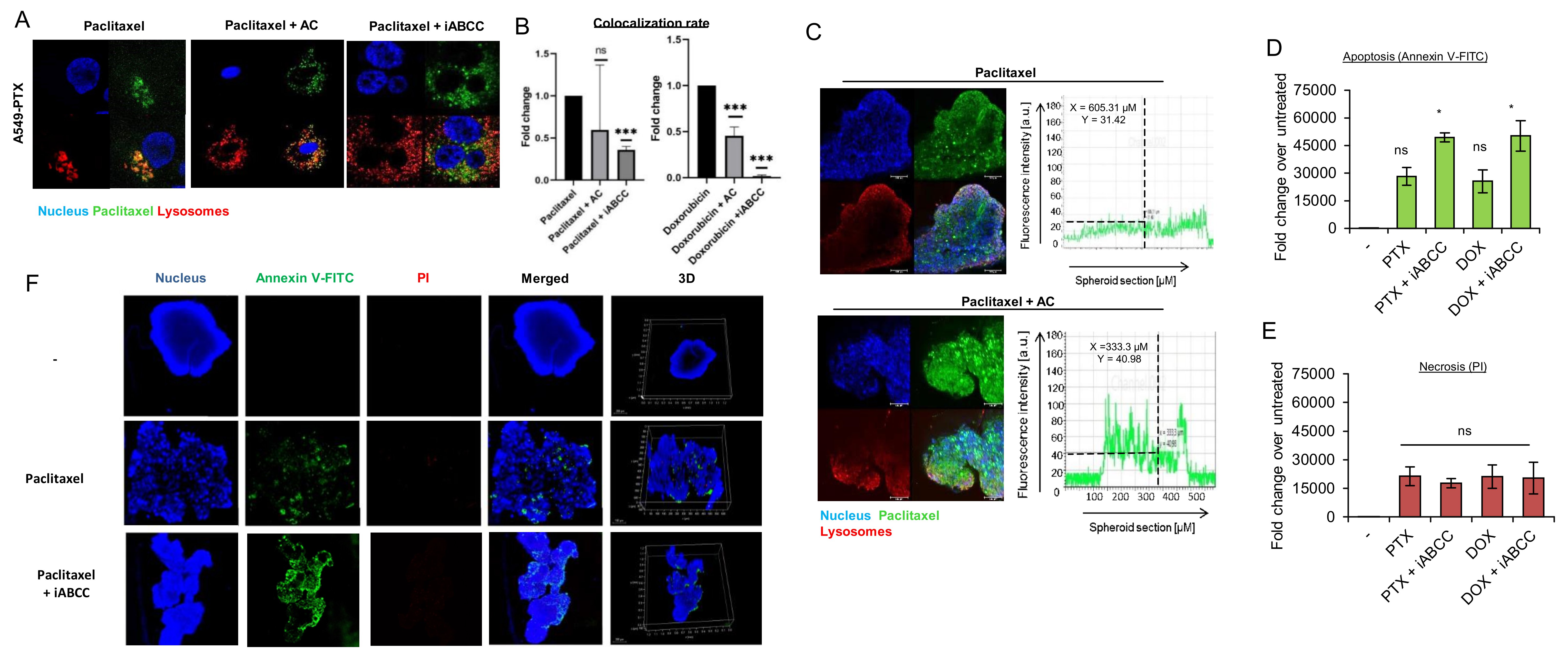
Fig. 3: Lysosomotrophic neutralizing agent prevents ABCC-dependent drug sequestration in lysosomes of paclitaxel-resistant cancer phenotypes. A) Comparison of lysosome and paclitaxel colocalization in paclitaxel-resistant A549 cell line after cell treatment with drug alone or in combination with lysosomotrophic neutralizing agent (ammonium chloride - AC; 25 mM) and ABCC inhibitor (MK-571 – iABCC; 100 µM), which were added for 2 h prior to Paclitaxel Oregon GreenTM 488 treatment (0.1 µM). Pictures were acquired with confocal microscope (TCS SP8, Leica Microsystems, Germany). Paclitaxel Oregon GreenTM 488 is marked red, DNA was stained with DAPI (blue) and lysosomes with LysoTrackerTM Deep Red (green). (B) The impact of ammonium chloride (AC) and MK571 (iABCC) on doxorubicin (0,05 µM) and paclitaxel (0.1 µM) colocalization with lysosomes was determined in arbitrary units (a.u.) with Leica Application Suite X. The difference between two means was tested with Student’s t-test or Mann Whitney test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001. (C) 4 week-old 3D PTX-resistant A549 were treated with Paclitaxel Oregon GreenTM 488 (green) alone for 48 h or in combination with ammonium chloride (25 mM; 2 h prior to paclitaxel), stained with LysoTrackerTM Deep Red (red) and DAPI (blue). Spheroid fluorescence was captured with confocal microscope (TCS SP8, Leica Microsystems, Germany). The fluorescence intensity plot at spheroid cross section was determined in arbitrary units (a.u.) with Leica Application Suite X. (D,E) The role of ABCC in cell protection against doxorubicin (0.05 μM; 48 h) and paclitaxel (0.1 μM; 48 h) -induced cytotoxicity in 3D culture of 3 week old A549-PTX spheroids was assayed by measurement the Annexin V (D) and propidium iodide. (E) iABCC was added as in (A). The fluorescence intensity was determined in arbitrary units (a.u.) with Leica Application Suite X. The difference between the variants was tested with one-way ANOVA and Tukey post-hoc test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001. (F) The role of ABCC in cell protection against paclitaxel-induced cytotoxicity (0.01 μM; 48 h) in 3D culture of 3 week old A549-PTX spheroids was assayed by spheroid triple staining with Annexin V (green), propidium iodide (red) and DAPI (blue) for 1 h. iABCC was added as in A. Images were acquired as in C. The green fluorescence intensity of entire spheroid corresponds to the extent of apoptosis in the culture, whereas intensity of red fluorescence to necrosis.
Deficiency of ABCC3, ABCC5 and ABCC10 prevents drug sequestration in lysosomes of paclitaxel-resistant cancer cells
To identify the possible role of particular ABCC family members in lysosomal drug sequestration, we made use of transient gene silencing and targeted mRNA of ABCC3, ABCC5 and ABCC10 with siRNA to substantially reduce their abundance in paclitaxel-resistant cells (Fig. S6). We considered whether these proteins contributed to regulation of the drug level inside cells. Either flow cytometry measurement of Paclitaxel Oregon Green 488 and doxorubicin fluorescence intensity inside whole cells (Fig. 4A) or confocal microcopy-based quantification of cell fluorescence (Fig. 4B, 4C) indicated that there was no sign of substantial alteration in cell fluorescence, with only one exception. The deficiency of ABCC3 in A549-PTX was followed by a statistically significant increase of the doxorubicin level inside cells when analyzed by flow cytometry. However, the silencing of ABCC3, ABCC5 or ABCC10 increased the drug level outside lysosomes (Fig. 4B), hence reducing drug colocalization with lysosomes (Fig. 4D). The most prominent reduction in doxorubicin-lysosome and Paclitaxel Oregon Green 488-lysosome co-occurrence was observed in ABCC3-deficient cells. This protein therefore seems to be largely responsible for the accumulation of fluorescently-labeled paclitaxel and anthracycline inside lysosomes.
To confirm the role of particular ABCC transporters in lysosomal drug sequestration, these organelles were isolated from paclitaxel-resistant ABCC3-, ABCC5- and ABCC10-deficient and proficient cells treated with Paclitaxel Oregon Green 488 and compared using lysosome fluorescence by flow cytometry. As shown in Fig. 4E and Fig. S7 and according to data from confocal imaging (Fig. 4B, 4C, Fig S8), the most striking reduction of fluorescently labeled paclitaxel accumulation was observed in ABCC3-deficient cells, where it dropped below 50%. The knock down of the other two proteins caused slight, mostly statistically insignificant changes with the exception of ABCC10 knock down in paclitaxel-resistant A549 cells, where the accumulation of Paclitaxel Oregon Green 488 was considerably reduced.
These results suggests that all three studied proteins may be important for lysosome-based drug accumulation and resistance, with ABCC3 emerging as a key protein involved in the active trapping of chemotherapeutics inside acidic organelles.
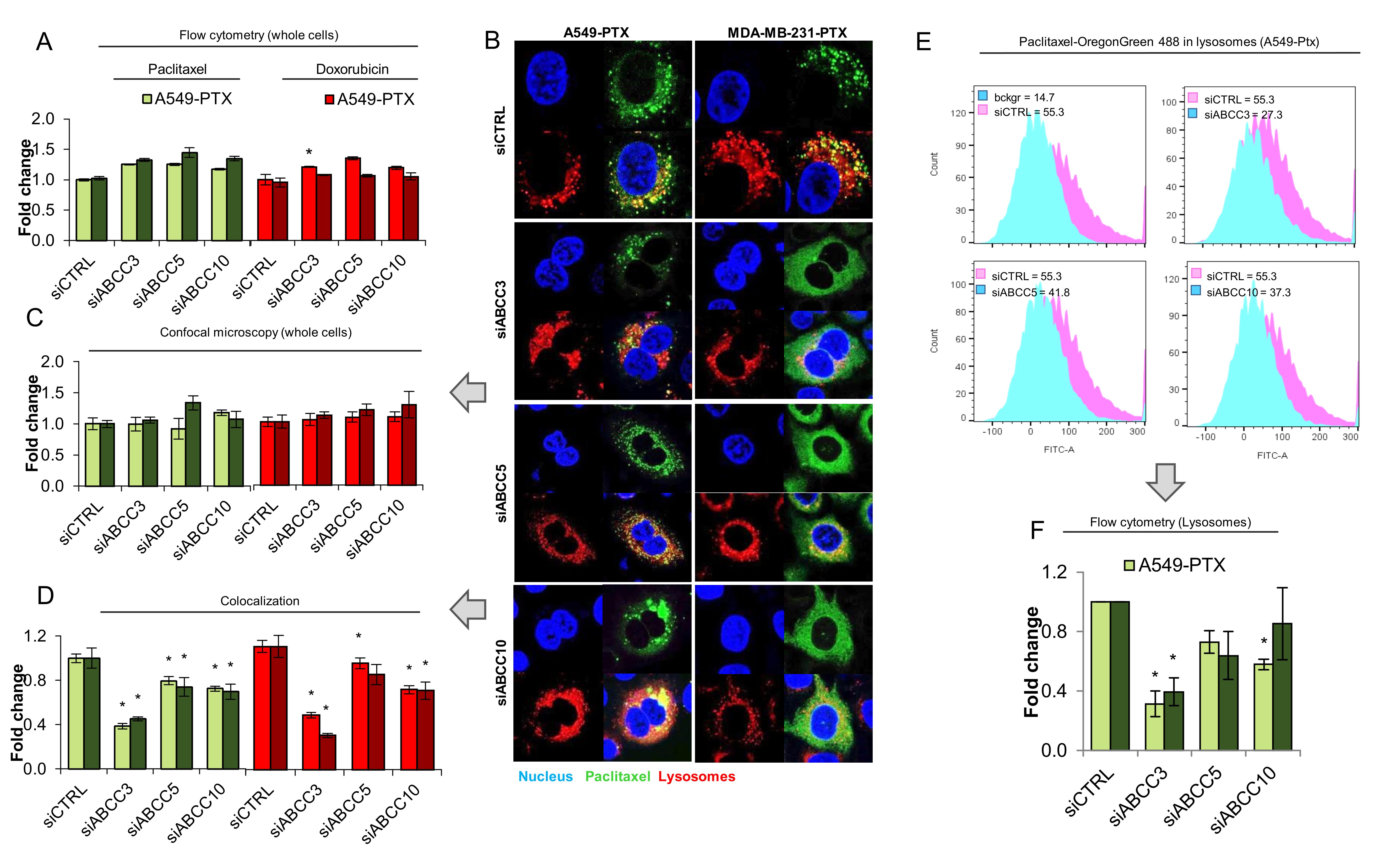
Fig. 4: Transient ABCC silencing prevents drug sequestration in lysosomes of paclitaxel-resistant cancer phenotypes. (A)Comparison of cell lysosomal uptake in paclitaxel-resistant A549 cell line and MDA-MB-231 after transfection with control siRNA and siRNA for ABCC3, ABCC5 and ABCC10. Paclitaxel (0.1 µM) and Doxorubicin (0.05 µM) was added 48h after transfection for 24h. The fluorescence intensity was measured by a flow cytometer LSR® II (Becton Dickinson) at ex: 470/em: 595 nm/nm for anthracyclines and ex: 496 nm/em: 524 nm for paclitaxel. Distribution of cells intensity was analysed in Flowing Software 2. (B,C,D) Comparison of paclitaxel distribution in paclitaxel-resistant A549 cell line (A) and MDA-MB-231 after transfection with control siRNA and siRNA for ABCC3, ABCC5 and ABCC10. Paclitaxel (0,1 µM) was added 48h after transfection for 24h. (B) Pictures were acquired with confocal microscope (TCS SP8, Leica Microsystems, Germany). Paclitaxel Oregon GreenTM 488 is marked green, DNA was stained with DAPI (blue) and lysosomes with LysoTrackerTM Deep Red (red). The fluorescence intensity, which corresponds to drug level inside cells (C) and colocalization between considered proteins and lysosomes (D) were determined in arbitrary units (a.u.) with Leica Application Suite X. The difference between the variants was tested with one-way ANOVA or Kruskal Wallis Test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001. (E,F) Comparison of paclitaxel lysosomal uptake in paclitaxel-resistant A549 cell line and MDA-MB-231 after transfection with control siRNA and siRNA for ABCC3, ABCC5 and ABCC10. Paclitaxel (0,01 µM) was added 48h after transfection for 24h. The fluorescence intensity was measured by flow cytometer LSR® II (Becton Dickinson) at em: 524 nm/ex: 496 nm. Distribution of lysosomes intensity was analysed in Flowing Software 2.
Lysosome-enriched ABCC transporters protect paclitaxel-resistant cancer cells from anticancer drugs
Since the impact of single ABCC silencing on doxorubicin and Paclitaxel Oregon Green 488 accumulation in lysosomes was incomplete, all three proteins were knocked down simultaneously and the drug accumulation and toxicity was compared for ABCC3/5/10-deficient and proficient cells. As expected, targeting mRNA of three ABCC transporters with siRNA lowered lysosomal sequestration of doxorubicin and fluorescently-labeled paclitaxel (Fig. 5A, 5B, Fig. S9A), and led to dramatic decrease in drug-lysosome colocalization, up to ~4% for doxorubicin in the paclitaxel-resistant A549 cells (Fig. 5C, Fig. S9B). Importantly, the knock down of ABCC3/ABCC5/ABCC10 increased the cytotoxicity of paclitaxel, doxorubicin, cisplatin and etoposide that vary in their acidity. A similar effect was also observed for the pan-ABCC inhibitor (Fig. 5D).
In a 3D culture of these cells, the knock down of all three ABCC transporters substantially enhanced the spheroid penetration by Paclitaxel Oregon Green 488, which was distributed approximately equally across the entire spheroid regardless of its depth, whereas in control cells the green fluorescence of the drug was prevalently observed in the outer spheroid layers (Fig. 5E). A similar profile of the difference between siCTRL and siABCC was also found in doxorubicin-treated 3D cell cultures (Fig. S9C). Furthermore, silencing of ABCC3/5/10 phenocopied the impact of iABCC and ammonium chloride regarding drug distribution to spheroids. This suggests that ABCC, enriched in lysosomes of paclitaxel-resistant cancer cells, is involved in the doxorubicin and Paclitaxel Oregon Green 488 trapping in the outer cell layers of the 3D culture and protects deeper spheroid layers from drug penetration. To test if drug retention in the peripheral part of spheroids by ABCC3, ABCC5 and ABCC10 reduces the cytotoxicity of doxorubicin and paclitaxel similarly to the pan-ABCC inhibitor, we compared the extent of apoptotic and necrotic cells between the control and ABCC3/5/10 knockdowns (Fig. 5F, 5G). Analysis of apoptotic marker - Annexin V intensity in a 3D culture of A549-PTX and MDA-MB-231 spheroids confirmed a more potent apoptotic induction by the drug in ABCC knockdown cells than transfection with control siRNA. A greater extent of necrosis was observed in cells after ABCC silencing and doxorubicin treatment (Fig. S10C-D), but it was not observed after ABCC silencing and paclitaxel exposure, in comparison with control siRNA (Fig. S10A-B). Interestingly, Annexin V and propidium iodide-stained spheroids showed a similar distribution of apoptotic cells in all spheroid layers after ABCC silencing in comparison with control siRNA in which apoptotic marker was located mainly in the outer parts of the spheroid (Fig. S10 A-D). These results suggest that drug entrapment by lysosomal ABCC3, ABCC5 and ABCC10 reduces drug cytotoxicity.
In summary, our results suggest that the lysosomal fractions of ABCC3, ABCC5 and ABCC10 may protect cells growing in 3D structures from doxorubicin and paclitaxel. Their impact on the anticancer efficacy of other chemotherapeutics that are actively transported across membranes by ABCC3, ABCC5 and ABCC10, is also likely, but requires further verification.
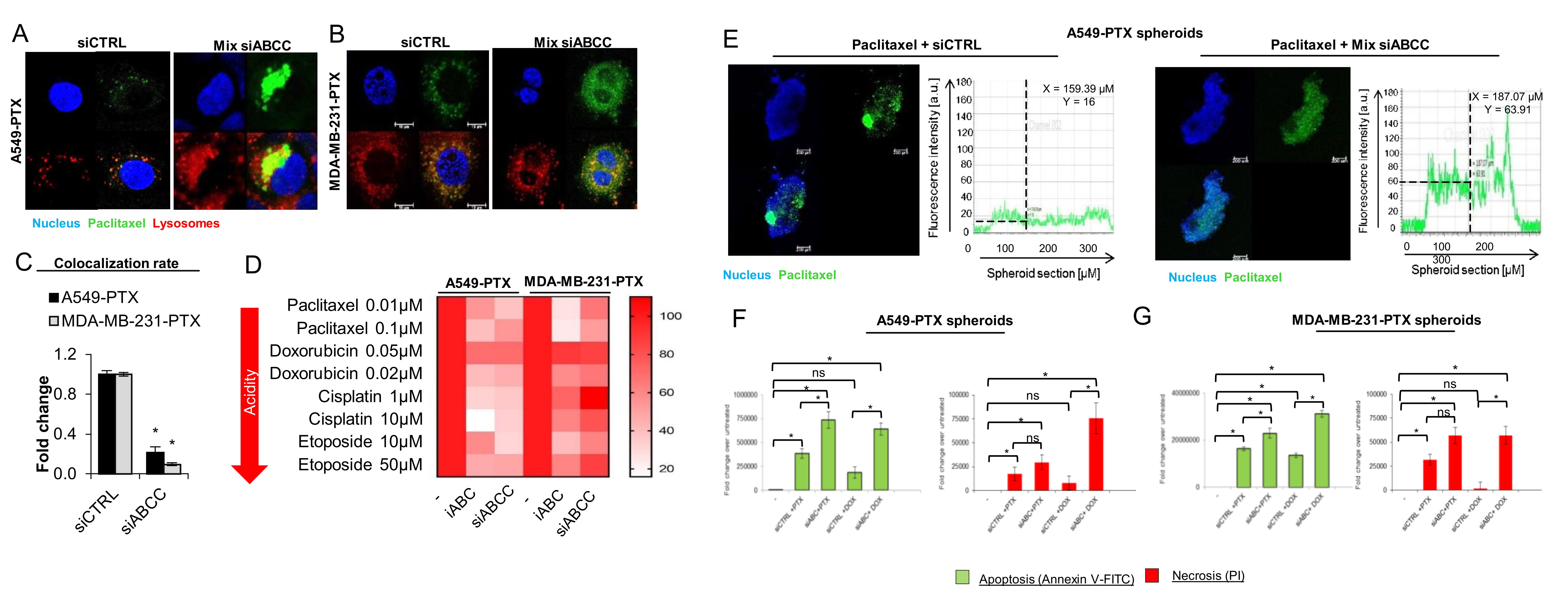
Fig. 5: Transient ABCC silencing increase cytotoxic effect of anticancer drugs. (A-C) The impact of simultaneous silencing with siABCC3, siABCC5 and siABCC10 on Paclitaxel Oregon Green 488 intracellular distribution was tested in paclitaxel-resistant A549 (A) and MDA-MB-231 (B) cells. Fluorescently labeled paclitaxel (green) was added to cells 72 h after their transfection with siRNA mixture (Mix siABCC) or with non-template control for another 24 h. Lysosomes were stained with LysoTrackerTM Deep Red (red) and DAPI (blue). (C) Colocalization of Paclitaxel Oregon Green 488-derived green and LysoTrackerTM-derived red fluorescence was quantified with Leica Application Suite X. Bars in Fig. show mean ± SD. The difference between two means was tested with Student’s t-test (Gaussian distribution values) or Mann Whitney test (non-Gaussian distribution values), and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001. (D) Cell metabolic activity that serves as a readout of cell viability was assayed with resazurin red. iABCC (MK-571; 25 μM) was administrated to the culture for 2 h prior to drugs, whereas cell transfection with the mixture of siABCC3, siABCC5 and siABCC10 was carried out 72 h prior to induction of cytotoxicity. For each tested chemotherapeutic, the viability of siCTRL or iABCC untreated cells was assumed as 100%. (E) 4 week-old 3D PTX-resistant A549 were transfected with mix of ABCC3, ABCC5 and ABCC10 or siRNA control 78h prior to treatment with paclitaxel (0.1 µM), stained with DAPI (blue). Paclitaxel Oregon Green 488 fluorescence was captured with confocal microscope (TCS SP8, Leica Microsystems, Germany). The fluorescence intensity plot at spheroid cross section was determined in arbitrary units (a.u.) with Leica Application Suite X. (F,G) The role of ABCC3, ABCC5 and ABCC10 in cell protection against doxorubicin (0.05 μM; 48 h) and paclitaxel (0.1 μM; 48 h) -induced cytotoxicity in 3D culture of 3 week old A549-PTX and MDA-MB-231-PTX spheroids was assayed by measurement the Annexin V (D) and propidium iodide (E). The drugs was added to cells 72 h after their transfection with siRNA mixture (Mix siABCC) or with non-template control. The fluorescence intensity was quantified with Leica Application Suite X. The difference between the variants was tested with one-way ANOVA or Kruskal Wallis Test, and statistically significant differences are marked with * when p<0.05 *, ** when p<0.01, *** when p<0.001.
Discussion
One of the most frequently referred mechanism of multidrug resistance is conferred by the overexpression of ABC transporters that actively transport chemotherapeutic drugs across intracellular and plasma membranes, thereby removing them out of the cells or sequestering them inside some cellular organelles and limiting their toxicity. A substantial body of evidence links declined cancer cell responsiveness to chemotherapeutics being accumulated in lysosomes, which increase in number due to the G2 arrest-dependent activation of TFEB transcription factor following the exposure of cancer cells to hydrophobic weak-base drugs such as doxorubicin, daunorubicin, mitoxantrone and symadex [42]. Some data including our results, also indicated enhanced lysosomal biogenesis in paclitaxel-treated cancer cells [15], which may be assigned to at least two mechanisms. Similarly, to the drugs listed above, paclitaxel triggered G2/M arrest regardless of the p53 mutation status in glioma cells [43]. Furthermore, lysosomal drug sequestration induced lysosomal stress and TFEB-mediated lysosomal biogenesis [44]. Additionally, it has been shown that stress factors in the tumor microenvironment induced drug resistance via ABCB1. This occurred through two mechanisms: rapid P-glycoprotein internalization via endocytosis of plasma membrane and redistribution via intracellular trafficking and hypoxia-inducible factor-1α expression which up-regulated ABCB1 expression and was accompanied by lysosomal biogenesis. As part of endocytosis, the ABCB1-containing plasma membrane buds inward to form an early endosome, which next forms a lysosome. During endocytosis, the catalytic active-site and ATP-binding domain of P-glycoprotein are still exposed to the cytosol and are enabled to pump substrates from the cytosol into lysosomes [45]. Lysosomal drug accumulation results in the translocation of lysosomes from the perinuclear zone towards the plasma membrane via movement on microtubule tracks. Following translocation to the plasma membrane, drug-loaded lysosomes fuse with the plasma membrane and release the drugs from the cell, evidenced by the increasing levels of the lysosomal enzyme cathepsin D in the extracellular environment [46]. Other transporters, such as ABCA2-3, ABCC1-2 and ABCG2 were shown to be recruited from the cell membrane surface via internalization into (early/late) endo- or lysosomes, which was demonstrated in colocalization studies with these ABC transporters and endo- or lysosomal markers. However, some ABC transporters (e.g., ABCA4, ABCA5, ABCB1, ABCB6, ABCB9, ABCC4–5, ABCD4, ABCG1 and ABCG4) have, in addition to their cellular membrane localization, a real subcellular destination rather than an intermediate [16]. Nothing is known at present about the molecular mechanism that sorts the ABCC family members such as ABCC1, ABCC2, ABCC3, ABCC5 and ABCC10 to lysosomes and the lysosomal membrane [22]. Although some differences in amino acid sequence and protein structure between ABCB1 and these ABCC transporters exist, they share similarity in functional domains allowing them to bind to ATP and substrates, and so be shuttled across membranes [23]. Liu-Kreyche and co-authors provided evidence for the presence of ABCC2, ABCC3 and ABCC4 in similar or even higher levels than ABCB1 in lysosomes of non-resistant HepG2, Hep3B2, H226, OVCAR3 and N87 cell lines, and ABCC3 was detected in all of them [22]. In our model of A549 and MDA-MB-231 cells the development of drug resistance was followed by the enrichment of ABCC3, ABCC5 and ABCC10 in lysosomes, but only ABCC5 transcription was considerably enhanced. This suggests that the redistribution of ABC transporters from plasma or from other intracellular compartment membranes to lysosomes rather than protein overexpression, plays a key role in lysosomal sequestration of at least doxorubicin and Paclitaxel Oregon Green 488. This conclusion can be possibly extended to other anticancer therapeutics, which are listed as substrates of ABC transporters, however the trapping of these therapeutics inside acidic organelles in multidrug resistant cells must be first experimentally confirmed. The origin of ABCC proteins that are enriched in lysosomal membrane of paclitaxel-resistant cell lines remains unknown. None of the three proteins were visibly present in the plasma membrane of resistant cells, in contrast to ABCC1. Hence, the indirect pathway of their lysosomal targeting seems unlikely, where after synthesis in the endoplasmic reticulum and passing through the Golgi apparatus, they are first sorted to the cell surface and from there, enter the endocytic pathway in response to cell treatment with doxorubicin or Paclitaxel Oregon Green 488. The plasma membrane localized ABCC1 did not move to the lysosomes during development of paclitaxel resistance, thereby further supporting the hypothesis based on the alternative route of subcellular trafficking of lysosomal transmembrane ABCC proteins. The variety of options include: classical and direct sorting of endoplasmic reticulum-derived proteins from the trans-Golgi network (TGN) to the endosomes and lysosomes that are mediated by adaptor protein complexes (AP1, AP2, AP3), or Golgi-localized, gamma-ear containing ADP-ribosylation factor binding (GGA) proteins, or non-classical pathways such as post-translational modifications (N-glycosylation and covalent lipid attachment), several amino acid motifs that do not conform to the canonical tyrosine and dileucine signals and association with other proteins [47]. Other ABCA, ABCB and ABCD subfamily members are considered to be lysosomal proteins. ABCB6 and ABCB9 are internalized to endolysosomes in clathrin-dependent fashion by an extended N-terminal domain (TMD0), which does not contain classical sorting determinants. ABCD4 traffics to the lysosomes only in association with LMBD1 (LMBR1 domain-containing protein 1), a lysosomal protein with nine putative transmembrane domains and classical tyrosine-based motif located in a cytosolic loop. ABCA2, ABCA3 and ABCA4 possess the trafficking signal motive xLxxKN (proximal post-Golgi compartment) that drives them to lysosomes [23]. This suggests that cell exposure to paclitaxel and other drugs which induce lysosome-mediated multidrug resistance [15, 17, 21], may alter the ABCC intracellular trafficking pathways without apparent increase in their expression. This aspect requires further investigation since the ability to retain multidrug resistance-associated ABC transporters outside lysosomes could improve the efficacy of chemotherapy. Another important issue refers to the functional impact of ABC transporters on drug sequestration in lysosomes and limiting their toxicity. Previous reports have focused mostly on P-glycoprotein with its involvement in multidrug resistance and colocalization with lysosomal LAMP2 protein in vinblastine- and paclitaxel-resistant (but not in non-resistant) cervical and ovary cancer cells, respectively [33]. Despite experimental evidence indicating P-glycoprotein incorporation into the membrane of cytoplasmic vesicles, the contribution of this enzyme in drug sequestration remains controversial. The enzyme remains active in intracellular compartments but its ER and Golgi localization is transient and lysosomal localization is less common, even while the lysosomal pathway plays a major role in the degradation of P-gp in cancer cells [23]. The silencing of ABCB1, with pharmacological inhibition of the enzyme with valspodar or elacridar and treatment with lysosomotropic weak bases (NH4Cl, chloroquine, or methylamine), caused relocalization of DOX from the lysosomes to DNA in the nucleus and sensitized only the Pgp-expressing cell to cytotoxic drugs. Accordingly, the paclitaxel-resistant breast and lung cancer cell lines were characterized by a striking enrichment of ABCC3, ABCC5 and ABCC10 in lysosomes, which was followed by the enhanced accumulation of doxorubicin and paclitaxel-OregonGreen in lysosomes of paclitaxel-resistant phenotypes. Though direct evidence is lacking regarding the functional expression of P-glycoprotein in the membrane of lysosomes. Regardless whether P-glycoprotein activity in lysosomes has been corroborated or not, there was no correlation shown between the patterns of the expression level of ABCB1 and the resistance to paclitaxel in human lung cancer cell lines. The colocalization between Oregon Green® 488 conjugated paclitaxel with LysoTracker® Red was more frequent in the RERF-LC-KJ cells, characterized by relatively low expression of ABCB1 when compared to other cell lines [15]. This leaves a gap and suggests the likely contribution of other anticancer drug transporters such as ABCC accumulating Oregon Green® 488 conjugated paclitaxel in the lysosomes of cells that are low in ABCB1. In our study ABCC3 emerged the most potent for trapping Paclitaxel Oregon Green 488 and doxorubicin in lysosomes, although confocal microscopy images and quantification of ABCC5 and ABCC10 colocalization with lysosomes confirmed their role in the lysosomal trapping of both studied compounds. The advantage of one transporter over the other in the sequestration of particular chemotherapeutics may be determined by their abundance in the lysosomal membrane as well as their substrate specificity. ABCC3 transports a variety of antineoplastic drugs such as etoposide, vincristine, methotrexate and sorafenib, whereas its overexpression has correlated to the sensitivity reduction of drugs, namely paclitaxel, docetaxel, gemcitabine, vinorelbine, cisplatin, doxorubicin and VP-16 [48]. The substrates of ABCC5 comprise nucleoside analogs, cyclic nucleosides, 6-mercaptopurine and thioguanine, 5-fluorouracil, cisplatin, methotrexate, PMEA, AZT, but also a wide range of fluorochromes [49]. Therefore, limited activity of ABCC5 in Paclitaxel Oregon Green 488 and the doxorubicin loading to lysosomes may result from a limited specificity towards the studied compounds. However, the impact of ABCC10 on sequestration of our drugs was lower than that of ABCC3 even though paclitaxel and doxorubicin are listed among ABCC10 substrates [50]. Our unpublished RNA-Seq data confirms the highest mRNA level of ABCC3 in paclitaxel-resistant A549 cells, and a 10-fold lower level of ABCC5 and ABCC10. Interestingly, the enrichment of ABCB1 in the lysosomes of paclitaxel-resistant cells did not have any substantial impact on ABCC3, ABCC5 and ABCC10-dependent drug trapping in lysosomes. As mentioned above, the abundance of particular proteins in the lysosomal membrane ensures their bona fide contribution to drug transport into the lysosome lumen. From our unpublished data, the mRNA level of ABCB1 was more than 10-fold lower than that of ABCC5 and ABCC10, and more than 50-fold lower than ABCC3 in both paclitaxel-resistant cell lines. Therefore, the role of ABCB1 in our cellular model may be of minor importance.
Cancer cells are characterized by a large heterogeneity in the expression of ABC transporters. mRNA profiling of all known human ABC genes and subsequent Western Blot analysis of their protein expression showed an overlap in the overexpression of ABCB1 and ABCC3 proteins only in the tested paclitaxel-resistant breast cancer sublines. Up-regulation of ABCG2 and ABCC4 proteins was observed only in paclitaxel-resistant SK-BR-3 cells and ABCB4 and ABCC2 in paclitaxel- resistant MCF-7 cells [11]. Due to the variability in transporter overexpression among tumors escalating during therapy, specific inhibition of particular transporters or their silencing, leads to the expected benefit in an unlikely way. In our study, pan-ABCC inhibitor – MK-571 with documented activity against ABCC1, ABCC2, ABCC3, ABCC5 and ABCC10 prevented doxorubicin and Paclitaxel Oregon Green 488 loading to lysosomes and enabled the toxicity of at least some chemotherapeutic drugs. Therefore, Pan-ABCC inhibitors emerge as encouraging candidates for pharmacological strategies to overcome multidrug resistance by exploiting lysosomes as subcellular targets in reversing drug sequestration resistance [44]. Many efforts have been made to interfere with the activity of ABC transporters and to reverse multidrug resistance. For example, P-glycoprotein, ABCC1 and BRCP inhibitors have been tested as chemo-sensitizers in clinical trials such as cyclosporine A and tariquidar. However, no useful therapeutic effects were shown due to high toxicity, drug-drug interaction and clinical trial design problems of this method [51]. New and safe ABCC transporter inhibitors capable of blocking multiple ABCC proteins for single or combinatorial patient treatment must be developed. Regardless of the overexpression profile or subcellular localization of particular ABCC proteins in a given tumor, pan-ABCC inhibitors could reduce drug efflux outside cells and limit drug sequestration in lysosomes. As mentioned previously, the relative level of ABCB1 expression and lysosome abundance, which is often elevated in drug-resistant cells, could predict the possible therapeutic success of pan-ABCC inhibitors. In particular, identification of lysosomes as essential players in drug resistance can lead to innovation of therapeutic approaches where targeting lysosomal function might improve response to chemotherapy [17].
In summary, the members of the ABCC subfamily of transmembrane transporters play a crucial role in lysosomal sequestration and hence the reduced toxicity of some anticancer drugs. The lysosomal trafficking of ABC proteins does not require their gene overexpression nor plasma membrane localization, typical for indirect protein sorting to the cell surface, to the endocytic pathway and hence, the lysosomal membrane. We provided further evidence that a passive influx of doxorubicin to lysosomes is facilitated by ABCC proteins, particularly ABCC3. Still, numerous questions remain open, including: why are these three proteins enriched in lysosomes? Why does ABCC1 remain in the plasma membrane upon ABC protein mobilization to lysosomes? Is the profile of the ABCC proteins sorted to lysosomes similar in other multidrug resistant cells?
Conclusion
In conclusion, our study shows that the lysosomal membrane of paclitaxel-resistant non-small cell lung cancer and triple-negative breast cancer is enriched in ABCC3, ABCC5 and ABCC1, contributing to doxorubicin and Paclitaxel Oregon Green 488 sequestration in lysosomes. Further studies are needed to confirm the role of the ABCC subfamily members in lysosome-mediated multidrug resistance, and their functional impact on other anticancer drug trapping. The identification of the mechanisms that cause a higher abundance of these particular ABCC proteins in lysosomes, other organelles and the plasma membrane, may provide new molecular targets for anticancer interventions.
Acknowledgements
Author Contributions
Conceptualization: A.R.; methodology: A.R. and K.G.; validation: K.G. and A.R.; investigation: K.G., S.M. and A.R; writing—original draft preparation: K.G and A.R.; writing—review and editing: K.G., S.M. and A.R.; supervision, A.R.; project administration, A.R.; funding acquisition, A.R.
Funding Sources
This research was funded by National Centre for Research and Development, grant number LIDER/22/0122/L-10/18/NCBR/2019.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Kampan NC, Madondo MT, McNally OM, Quinn M, Plebanski M: Paclitaxel and Its Evolving Role in the Management of Ovarian Cancer. Mulik RS, editor. Biomed Res Int 2015;2015:413076.
https://doi.org/10.1155/2015/413076 |
| 2 | Zang F, Rao Y, Zhu X, Wu Z, Jiang H: Shikonin suppresses NEAT1 and Akt signaling in treating paclitaxel-resistant non-small cell of lung cancer. Mol Med 2020;26:28.
https://doi.org/10.1186/s10020-020-00152-8 |
| 3 | Pu J, Shen J, Zhong Z, Yanling M, Gao J: KANK1 regulates paclitaxel resistance in lung adenocarcinoma A549 cells. Artif cells, nanomedicine, Biotechnol 2020;48:639-647.
https://doi.org/10.1080/21691401.2020.1728287 |
| 4 | Ge J, Zuo W, Chen Y, Shao Z, Yu K: The advance of adjuvant treatment for triple-negative breast cancer. Cancer Biol Med 2021;19:187-201.
https://doi.org/10.20892/j.issn.2095-3941.2020.0752 |
| 5 | Maloney SM, Hoover CA, Morejon-Lasso L V, Prosperi JR: Mechanisms of Taxane Resistance. Cancers 2020;12:11.
https://doi.org/10.3390/cancers12113323 |
| 6 | Škubník J, Pavlíčková V, Ruml T, Rimpelová S: Current Perspectives on Taxanes: Focus on Their Bioactivity, Delivery and Combination Therapy. Plants 2021;10:3.
https://doi.org/10.3390/plants10030569 |
| 7 | Pan S-T, Li Z-L, He Z-X, Qiu J-X, Zhou S-F: Molecular mechanisms for tumour resistance to chemotherapy. Clin Exp Pharmacol Physiol 2016;43:723-737.
https://doi.org/10.1111/1440-1681.12581 |
| 8 | Gao L, Zhao P, Li Y, Yang D, Hu P, Li L, et al.: Reversal of P-glycoprotein-mediated multidrug resistance by novel curcumin analogues in paclitaxel-resistant human breast cancer cells. Biochem Cell Biol 2020;98:484-91.
https://doi.org/10.1139/bcb-2019-0377 |
| 9 | A Phase I Study of Infusional Paclitaxel With the P-Glycoprotein Antagonist PSC 833 - Full Text View - ClinicalTrials.gov [Internet] [cited 2023 Apr 7].. Available from: https://clinicaltrials.gov/ct2/show/NCT00001383?cond=paclitaxel+ABCB1&draw=2&rank=2
|
| 10 | Paclitaxel and CBT-1(Registered Trademark) to Treat Solid Tumors - Full Text View - ClinicalTrials.gov [Internet] [cited 2023 Apr 7].. Available from: https://clinicaltrials.gov/ct2/show/NCT00972205?cond=paclitaxel+ABCB1&draw=2&rank=5
|
| 11 | Němcová-Fürstová V, Kopperová D, Balušíková K, Ehrlichová M, Brynychová V, Václavíková R, et al.: Characterization of acquired paclitaxel resistance of breast cancer cells and involvement of ABC transporters. Toxicol Appl Pharmacol 2016;310:215-228.
https://doi.org/10.1016/j.taap.2016.09.020 |
| 12 | Hou Y, Zhu Q, Li Z, Peng Y, Yu X, Yuan B, et al.: The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis 2017;8:e2659-e2659.
https://doi.org/10.1038/cddis.2017.53 |
| 13 | Stefan SM, Wiese M: Small-molecule inhibitors of multidrug resistance-associated protein 1 and related processes: A historic approach and recent advances. Med Res Rev 2019;1;39:176-264.
https://doi.org/10.1002/med.21510 |
| 14 | Alfarouk KO, Stock CM, Taylor S, Walsh M, Muddathir AK, Verduzco D, et al.: Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell Int [Internet]. 2015 Jul 15 [cited 2023 Apr 7];15:1-13.
https://doi.org/10.1186/s12935-015-0221-1 |
| 15 | Shimomura M, Yaoi T, Itoh K, Kato D, Terauchi K, Shimada J, et al.: Drug resistance to paclitaxel is not only associated with ABCB1 mRNA expression but also with drug accumulation in intracellular compartments in human lung cancer cell lines. Int J Oncol 2012;40:995-1004.
https://doi.org/10.3892/ijo.2011.1297 |
| 16 | Stefan K, Wen Leck LY, Namasivayam V, Bascuñana P, Huang ML-H, Riss PJ, et al.: Vesicular ATP-binding cassette transporters in human disease: relevant aspects of their organization for future drug development. Futur Drug Discov 2020;2:FDD51.
https://doi.org/10.4155/fdd-2020-0025 |
| 17 | Geisslinger F, Müller M, Vollmar AM, Bartel K: Targeting Lysosomes in Cancer as Promising Strategy to Overcome Chemoresistance-A Mini Review Front Oncol 2020;9;10:1156.
https://doi.org/10.3389/fonc.2020.01156 |
| 18 | Gong Y, Duvvuri M, Krise JP: Separate roles for the Golgi apparatus and lysosomes in the sequestration of drugs in the multidrug-resistant human leukemic cell line HL-60. J Biol Chem. 2003;278:50234-50239.
https://doi.org/10.1074/jbc.M306606200 |
| 19 | Stefan SM, Jansson PJ, Kalinowski DS, Anjum R, Dharmasivam M, Richardson DR: The growing evidence for targeting P-glycoprotein in lysosomes to overcome resistance. Future Med Chem 2020;12:473-477
https://doi.org/10.4155/fmc-2019-0350 |
| 20 | Bakos É, Homolya :. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1). Pflugers Arch Eur J Physiol 2007;23;453:621-641.
https://doi.org/10.1007/s00424-006-0160-8 |
| 21 | Gericke B, Wienböker I, Brandes G, Löscher W: Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver. Cells 2022;11:1556.
https://doi.org/10.3390/cells11091556 |
| 22 | Liu-Kreyche P, Shen H, Marino AM, Iyer RA, Humphreys WG, Lai Y: Lysosomal P-gp-MDR1 Confers Drug Resistance of Brentuximab Vedotin and Its Cytotoxic Payload Monomethyl Auristatin E in Tumor Cells. Front Pharmacol 2019;10:749.
https://doi.org/10.3389/fphar.2019.00749 |
| 23 | Szakacs G, Abele R: An inventory of lysosomal ABC transporters. FEBS Lett 2020;1;594:3965-3985.
https://doi.org/10.1002/1873-3468.13967 |
| 24 | Lee MM, Gao Z, Peterson BR: Synthesis of a Fluorescent Analogue of Paclitaxel that Selectively Binds Microtubules and Sensitively Detects Efflux by P-Glycoprotein. Angew Chem Int Ed Engl 2017;56:6927.
https://doi.org/10.1002/anie.201703298 |
| 25 | Stage TB, Bergmann TK, Kroetz DL: Clinical Pharmacokinetics of Paclitaxel Monotherapy: An Updated Literature Review. Clin Pharmacokinet 2018;57:7-19.
https://doi.org/10.1007/s40262-017-0563-z |
| 26 | Sousa C, Sá e Melo T, Gèze M, Gaullier J-M, Mazière JC, Santus R: Solvent Polarity and pH Effects on the Spectroscopic Properties of Neutral Red: Application to Lysosomal Microenvironment Probing in Living Cells. Photochem Photobiol 1996;63:601-607.
https://doi.org/10.1111/j.1751-1097.1996.tb05662.x |
| 27 | Sobczak M, Strachowska M, Gronkowska K, Robaszkiewicz A: Activation of ABCC Genes by Cisplatin Depends on the CoREST Occurrence at Their Promoters in A549 and MDA-MB-231 Cell Lines. Cancers (Basel) 2022;14:894.
https://doi.org/10.3390/cancers14040894 |
| 28 | FITC Annexin V Apoptosis Detection Kit with PI [cited 2023 Apr 21].; Available from: www.biolegend.com
|
| 29 | Guo W, Dong W, Li M, Shen Y: Mitochondria P-glycoprotein confers paclitaxel resistance on ovarian cancer cells. Onco Targets Ther 2019;12:3881-3891.
https://doi.org/10.2147/OTT.S193433 |
| 30 | Chung WM, Ho YP, Chang WC, Dai YC, Chen L, Hung YC, et al.: Increase Paclitaxel Sensitivity to Better Suppress Serous Epithelial Ovarian Cancer via Ablating Androgen Receptor/Aryl Hydrocarbon Receptor-ABCG2 Axis. Cancers (Basel) 2019;11:4.
https://doi.org/10.3390/cancers11040463 |
| 31 | Reshma PL, Unnikrishnan BS, Preethi GU, Syama HP, Archana MG, Remya K, et al.: Overcoming drug-resistance in lung cancer cells by paclitaxel loaded galactoxyloglucan nanoparticles. Int J Biol Macromol 2019;136:266-274.
https://doi.org/10.1016/j.ijbiomac.2019.06.075 |
| 32 | Lee MM, Gao Z, Peterson BR: Synthesis of a Fluorescent Analogue of Paclitaxel That Selectively Binds Microtubules and Sensitively Detects Efflux by P-Glycoprotein. Angew Chem Int Ed Engl 2017;56:6927-6931.
https://doi.org/10.1002/anie.201703298 |
| 33 | Yamagishi T, Sahni S, Sharp DM, Arvind A, Jansson PJ, Richardson DR: P-glycoprotein mediates drug resistance via a novel mechanism involving lysosomal sequestration. J Biol Chem 2013;288:31761-31771.
https://doi.org/10.1074/jbc.M113.514091 |
| 34 | Ruiz-López E, Jovčevska I, González-Gómez R, Tejero H, Al-Shahrour F, Muyldermans S, et al.: Nanobodies targeting ABCC3 for immunotargeted applications in glioblastoma. Sci Rep [Internet] 2022;12:22581.
https://doi.org/10.1038/s41598-022-27161-3 |
| 35 | Chen X-Y, Yang Y, Wang J-Q, Wu Z-X, Li J, Chen Z-S: Overexpression of ABCC1 Confers Drug Resistance to Betulin. Front Oncol 2021;11:640656.
https://doi.org/10.3389/fonc.2021.640656 |
| 36 | Barrington RD, Needs PW, Williamson G, Kroon PA: MK571 inhibits phase-2 conjugation of flavonols by Caco-2/TC7 cells, but does not specifically inhibit their apical efflux. Biochem Pharmacol 2015;95:193.
https://doi.org/10.1016/j.bcp.2015.03.005 |
| 37 | Pellegatta S, Di Ianni N, Pessina S, Paterra R, Anghileri E, Eoli M, et al.: ABCC3 Expressed by CD56dim CD16+ NK Cells Predicts Response in Glioblastoma Patients Treated with Combined Chemotherapy and Dendritic Cell Immunotherapy. Int J Mol Sci 2019;20:5886.
https://doi.org/10.3390/ijms20235886 |
| 38 | Döring H, Kreutzer D, Ritter C, Hilgeroth A: molecules Discovery of Novel Symmetrical 1, 4-Dihydropyridines as Inhibitors of Multidrug-Resistant Protein (MRP4) Efflux Pump for Anticancer Therapy. Molecules 2020;26:18.
https://doi.org/10.3390/molecules26010018 |
| 39 | Huang W, Chen K, Lu Y, Zhang D, Cheng Y, Li L, et al.: ABCC5 facilitates the acquired resistance of sorafenib through the inhibition of SLC7A11-induced ferroptosis in hepatocellular carcinoma. Neoplasia 2021;23:1227-1239.
https://doi.org/10.1016/j.neo.2021.11.002 |
| 40 | Kathawala RJ, Wang YJ, Ashby CR, Chen ZS: Recent advances regarding the role of ABC subfamily C member 10 (ABCC10) in the efflux of antitumor drugs. Chin J Cancer 2014;33:223.
https://doi.org/10.5732/cjc.013.10122 |
| 41 | Howe K, Gibson GG, Coleman T, Plant N: In silico and in vitro modeling of hepatocyte drug transport processes: importance of ABCC2 expression levels in the disposition of carboxydichlorofluroscein. Drug Metab Dispos 2009;37:391-399.
https://doi.org/10.1124/dmd.108.022921 |
| 42 | Mlejnek P, Havlasek J, Pastvova N, Dolezel P, Dostalova K: Lysosomal sequestration of weak base drugs, lysosomal biogenesis, and cell cycle alteration. Biomed Pharmacother 2022;153:113328.
https://doi.org/10.1016/j.biopha.2022.113328 |
| 43 | Han BI, Lee M: Paclitaxel-induced G2/m arrest via different mechanism of actions in glioma cell lines with differing p53 mutational status. Int J Pharmacol 2016;12:19-27.
https://doi.org/10.3923/ijp.2016.19.27 |
| 44 | Halaby R.: Influence of lysosomal sequestration on multidrug resistance in cancer cells. Cancer drug Resist (Alhambra, Calif) 2019;2:31-42.
https://doi.org/10.20517/cdr.2018.23 |
| 45 | Al-Akra L, Bae DH, Sahni S, Huang MLH, Park KC, Lane DJR, et al.: Tumor stressors induce two mechanisms of intracellular P-glycoprotein-mediated resistance that are overcome by lysosomal-targeted thiosemicarbazones. J Biol Chem 2018;293:3562.
https://doi.org/10.1074/jbc.M116.772699 |
| 46 | Zhitomirsky B, Assaraf YG: Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget 2017;8:45117.
https://doi.org/10.18632/oncotarget.15155 |
| 47 | Staudt C, Puissant E, Boonen M, Sansebastiano G-P Di, Gaballo A. Subcellular Trafficking of Mammalian Lysosomal Proteins: An Extended View. Int J Mol Sci 2016;18(1):47.
https://doi.org/10.3390/ijms18010047 |
| 48 | Ramírez-Cosmes A, Reyes-Jiménez E, Zertuche-Martínez C, Hernández-Hernández CA, García-Román R, Romero-Díaz RI, et al.: The implications of ABCC3 in cancer drug resistance: can we use it as a therapeutic target? Am J Cancer Res 2021;11:4127.
|
| 49 | Jungsuwadee P, Vore ME: Efflux Transporters. Compr Toxicol Second Ed 2010;4:557-601.
https://doi.org/10.1016/B978-0-08-046884-6.00426-7 |
| 50 | Abbasifarid E, Sajjadi-Jazi SM, Beheshtian M, Samimi H, Larijani B, Haghpanah V. The Role of ATP-Binding Cassette Transporters in the Chemoresistance of Anaplastic Thyroid Cancer: A Systematic Review. Endocrinology 2019;160(8):2015-23.
https://doi.org/10.1210/en.2019-00241 |
| 51 | Xiao H, Zheng Y, Ma L, Tian L, Sun Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front Pharmacol 2021;12:705.
https://doi.org/10.3389/fphar.2021.648407 |