Emerging Roles of SPT5 in Transcription
Keywords
Abstract
Introduction
Transcription is a highly orchestrated event divided into three phases: initiation, elongation, and termination [1][2][3][4]. The RNAP II interacts with several transcription and accessory factors during the above phases. These factors perform functions ranging from transcription to pre-mRNA processing [5]. The interaction of proteins with the RNAP II is dynamic and is coordinated spatially and temporally with the transcription machinery driving signals from intrinsic and extrinsic cellular signalling. Different proteins involved in different phases of transcription associate with the RNAP in a transitional manner. However, a few of them remain associated throughout the process. SPT5/SUPT5H is amongst some of the few proteins that remain associated with the RNAP throughout the transcription process.
SPT5/SUPT5H is one of the few universally conserved proteins across all three domains of life. Suggestive of the essential function it codes for even before the different life forms diverged. Our understanding of proteins and their role has increased in recent years, due to molecular, biophysical, and structural advances in proteins. In this review, we describe the Suppressor of Ty homolog-5 (SPT5/SUPT5H) as one of the few proteins that remain attached to the RNAP II and RNAP I from initiation to termination.
Discovery
SPT5/SUPT5H is a nuclear protein that regulates the transcription machinery of the cells. It is among the few proteins that are conserved across all three domains of life, suggesting its indispensability [6]. In bacteria, the SPT5 homolog is called NusG and functions as a monomer [7]. However, in archaea and eukaryotes, it functions as a heterodimer where it forms complexes with SPT4, another protein isolated in the same genetic screens and regulates the transcription [8]. SPT5 together with SPT4 forms a heterodimeric complex referred to as the DRB sensitivity-inducing factor (DSIF) that plays a role in regulating transcription [9][10][11].
SUPT5H is the human homolog of the yeast SPT5 protein. It was discovered in the yeast mutant screens as the mutant repressing mutations caused by the insertion of Ty element (His4-912δ) in the 5’noncoding region of the His4 gene in the Saccharomyces cerevisiae genetic screens [12][13]. Since the gene family was identified as suppressing the Ty element, the family was named Spt [9] [14]. Several other studies reported similar suppression of multiple histone mutations. Chromosome 13 codes for SPT5 in S. cerevisiae, while its human homolog is coded by chromosome 19[15]. SPT5 plays an essential role in transcription and regulates it in various steps, from promoter-proximal pausing to elongation and termination of RNAPII [16][17][18].
Structure
Structurally, SPT5 can be divided into five broad regions, each having a unique function. These include the N-terminal acidic region, a NusG N terminal (NGN) domain, multiple Kyprides-Ouzounis-Woese (KOW) domains, a C-terminal region (CTR) heptapeptide repetitive motif and the C-terminal [19].
The N-terminal region is a 150-170 amino acid long region composed of acidic amino acids and containing the nuclear localization signal at the 66th position. The signal is essential for the nuclear localization of the protein since its removal leads to the cytoplasmic localization of the protein [20]. A recent study by Park et al. highlighted the importance of the SPT5 N-terminal domain for the control of divergent antisense transcription at the transcription start site (TSS) [21]. For the first time, the study highlights the importance of N-terminal phosphorylation event of the SPT5 by the ZWC complex in the regulation of antisense transcription[22].
Adjacent to the N-terminal region is the NGN domain of the SPT5. It is the most conserved domain of the protein, remaining conserved throughout evolution in all three domains of life over the course of evolution [23]. The NGN domain harbours the site required for interaction with the RNAP II and SPT4 to form the DSIF complex [24][25]. The domain interacts with both the nascent RNA and the non-coding strand of the DNA. The hydrophobic concave depression on the surface of SPT5 interacts with the tip of the clamped coil domain of the RNAP II [26]. This interacting region remains largely intact in all domains of life (Fig 1A, B) . In archaea, alanine-4, tyrosine-42, and leucine-44 line in these hydrophobic pockets. Mutagenesis studies revealed that mutation of these residues significantly alters the binding to the RNAP.
Furthermore, replacing the ten residues of the clamp-coil motif with the polyglycine abolished the ability of RNAP II to interact with the NGN domain, thereby establishing conserved complementary binding. The NGN domain binds to the above DNA-RNA in the RNAP active site, thus locking the cleft for the nucleotide, resulting in increased elongation and decreased disassociation [26]. Furthermore, the negatively charged DNA also interacts with the positively charged surface of the Spt5, preventing the collapse of the transcription bubble. The NGN domain alters RNAP activity by altering the interaction with the clamp domain, thereby allosterically regulating the polymerase.
Linked to the NGN domain is the C-terminal KOW domain [27]. It is the other motif of SPT5, which remains relatively conserved across different domains. However, the number of KOW domains varies in each domain of life. The bacteria and archaea have a single KOW domain; however, comparatively, eukaryotes have multiple KOW domains ranging from four to six (Fig. 1A) [28][29][30]. The KOW domain is essential for survival since deletion leads to survival defects. KOW deletion mutants in the yeast showed severe growth defects.[31] The KOW domains of SPT5 adopt a fold similar to the β barrel of the Tudor domain, which known to interact with nucleic acid. The Tudor domain is structurally compact (~50 amino acids) and, facilitates protein-protein and protein-nucleic acid interactions [32]. In addition to the interactions with the nucleic acid, KOW domains of SPT5 could also be of essential importance for the coupling of the transcriptional modifications and the RNAP (Fig. 1C).
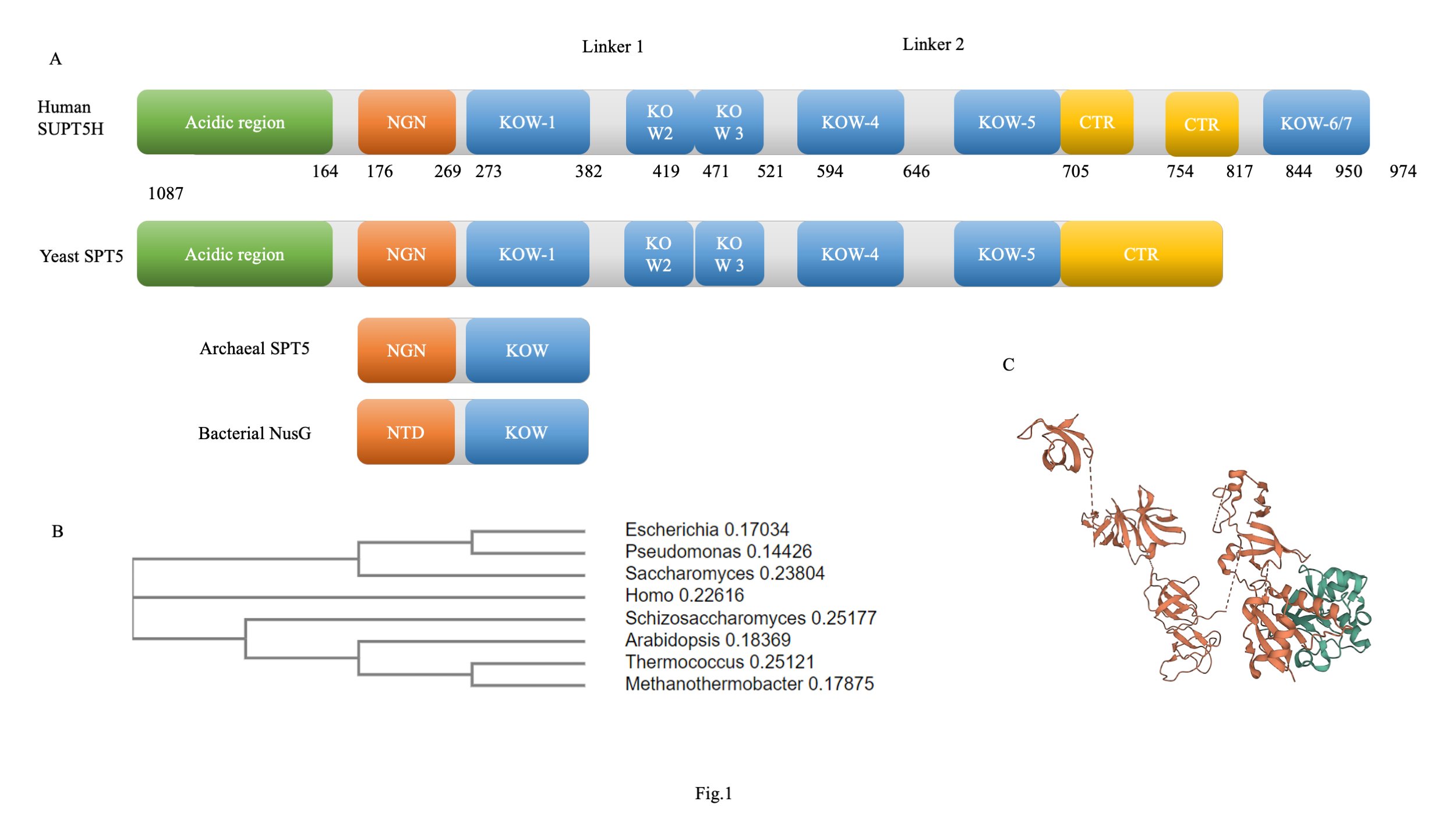
Fig. 1: SPT5 organization in different domains of life. (A) Domain organization of SPT5-like proteins present in Bacteria, Archaea, and Eukaryotes modified from Decker et al. Bacterial NusG protein and archaeal SPT5 proteins are comparatively small, consisting of either an N-terminal domain (NTD) or a NusG N-terminal domain (NGN) and a single KOW domain. In contrast, yeast SPT5 consists of an additional N-terminal acidic region followed by an NGN domain, 1-5 KOW domains, and a C-terminal repeat (CTR). Human SUPT5H consists of all the elements on the yeast SPT5 and an additional CTR followed by two additional KOW domains. Domain boundaries are drawn to proportionally according to their polypeptide lengths. (B) A cladogram showing evolutionary relatedness between different SPT5/NusG belonging to the different domains of life. (C) Structure of SPT5 (orange) bound to SPT4 (green) at transcription paused state. Other entities removed for better understanding (PDB ID: 6GML)
In eukaryotes, the first KOW domain folds together with the linker 1, the polypeptide sequence linking KOW1 to KOW2. KOW1-Linker1 (K1L1) folds into a bilobed rigid structure that serves as a DNA clamp. The N-terminal lobe is formed by the K1, which resembles the β barrel of the Tudor domain, along with some C-terminal sequences of L1 that form the final β strand of the β sheets. In contrast, the C-terminal lobe is formed by inserting sequences in the Tudor barrel. K1 and L1 have highly conserved interface residues in all eukaryotes, stabilized by five hydrogen bonds, van der Wall interaction, and a cation-π electron interaction [31]. High-resolution X-ray structures revealed a positively charged patch in K1LI that binds to the nucleic acid in a sequence-independent manner. In vitro studies showed that K1L1 prefers dsDNA over ssDNA and ssRNA. The function of KIL1 is complementary to the SPT4, which is also positively charged. Double mutation analysis of KIL1 and Spt4 caused severe growth arrest, which was rescued in the presence of wild-type Spt4, confirming the functional overlap [31]. K1LI is followed by the KOW2-3 tandem domain, having two Tudor domain β barrel structures separated by a single residue in the KOWX-4-KOW5 domain [33]. KOWX refers to pre-KOW4 region that forms the RNA clamp domain with KOW4-KOW5. KOWX-4 is a positively charged region that contacts the negatively charged RNA coming out of the ternary complex and blocks it from the action of RNAse. The KOW4-KOW5 linker features an essential phosphorylation site at S666 responsible for the productive elongation of the transcription complex. Acute loss or substitution at S666 results in proteasomal degradation to impaired pause release [34].
In addition to all of the mentioned domains, which are common to all eukaryotes, metazoans are characterized by an additional KOW domain at the end of the C-terminal repeat, i.e. KOW6-7 [35]. These domains are essential for maintaining SPT5’s negative regulatory role, as mutations result in defects in negative regulatory function, resulting in compromised growth [36].
Adjacent to the KOW domain lies the C-terminal repeat, which contains one to two repeating heptapeptide domains essential for transcription elongation [19][37]. The CTR mutant SPT5 is defective in transcription induction and causes lethality in the diploid state in S.cerevisiae; however, loss of CTR did not have such a lethal effect on inflammatory response genes in humans apart from slow growth [38][39]. CTR is the phosphorylation site for positive transcription elongation factor b (P-TEFb), which is responsible for phosphorylation of RNAP II CTD and recruitment of multiple elongation factors [40]. In addition, the CTR is the landing platform for several other proteins involved in the various transcription-coupled activities such as 5’capping, phosphorylation, 3’processing, and many others [41].
The C-terminal repeat terminates into the C-terminal domain; much is unknown. However, together with the CTR, it is thought to play a role in recruitment of proteins such as Paf-1, which is responsible for transcription elongation [42].
Biological Functions of SPT5
As a part of the DSIF complex, SPT5 associates RNAP II and regulates promoter-proximal pausing (Fig. 2A,B) [43]. The complex makes multiple contacts with the RNAP II around the active cleft and exit channel, thereby regulating its moment. In addition, several studies reported that SPT5 shares the same site as the transcription factors TFIIB, TFIIE, and TFIIF (Fig. 2C). In a validation study, it was suggested that the spt5-spt4 complex replaces the TFIIE, resulting in the promoter clearance of the pre-elongation complex (Fig. 2D) [44]. Therefore, it is evident that binding of SPT5 to the complex stabilizes the stalled complex. The stabilization of the stalled complex is further enhanced by the interaction of SPT5 with a protein known as the negative elongation factor (NELF)[45]. NELF binding alters the DNA-RNA hybrid in the transcription complex and impairs the NTP binding site, leaving the transcription complex in the paused state (Fig. 2E) [46][47]. The paused state is overcome upon the binding of P-TEFb to the complex [48]. P-TEFb is a heterodimeric complex of cyclin T1 and Cdk9 [49]. P-TEFb phosphorylates the SPT5, primarily at the two phosphorylation hot spot loci: the KOW4-KOW5 linker region and the C- terminal repeat domain [50]. Phosphorylation at the above site results in the release of NELF, resulting in transition from paused elongation complex to the active elongation complex. Removal of NELF alters the conformation, allowing entry of the nucleotides and progression of the transcription complexes (Fig. 2F) [51]. In addition to regulating the transcription, SPT5 ensures a constant rate of transcription and uniform distribution of RNAP II across the gene. In the absence of SPT5, a reduction in RNAP II levels and accumulation at the 5’ end of the gene occurs, further validated by the reduced level of mRNA post-SPT5 inhibition. A recent study by Aoi et al. reported an acute loss of SPT5 that led to the degradation of RPB1 of the RNAP II by the action of Cullin 3, VCP/p97 and CDK9 complex [52]. In addition, SPT5 was found to play an essential role in repressing antisense transcription. SPT5 inhibits two classes of antisense transcription, including convergent transcription downstream of the TSS, and the divergent transcription, upstream of the TSS [53][54].
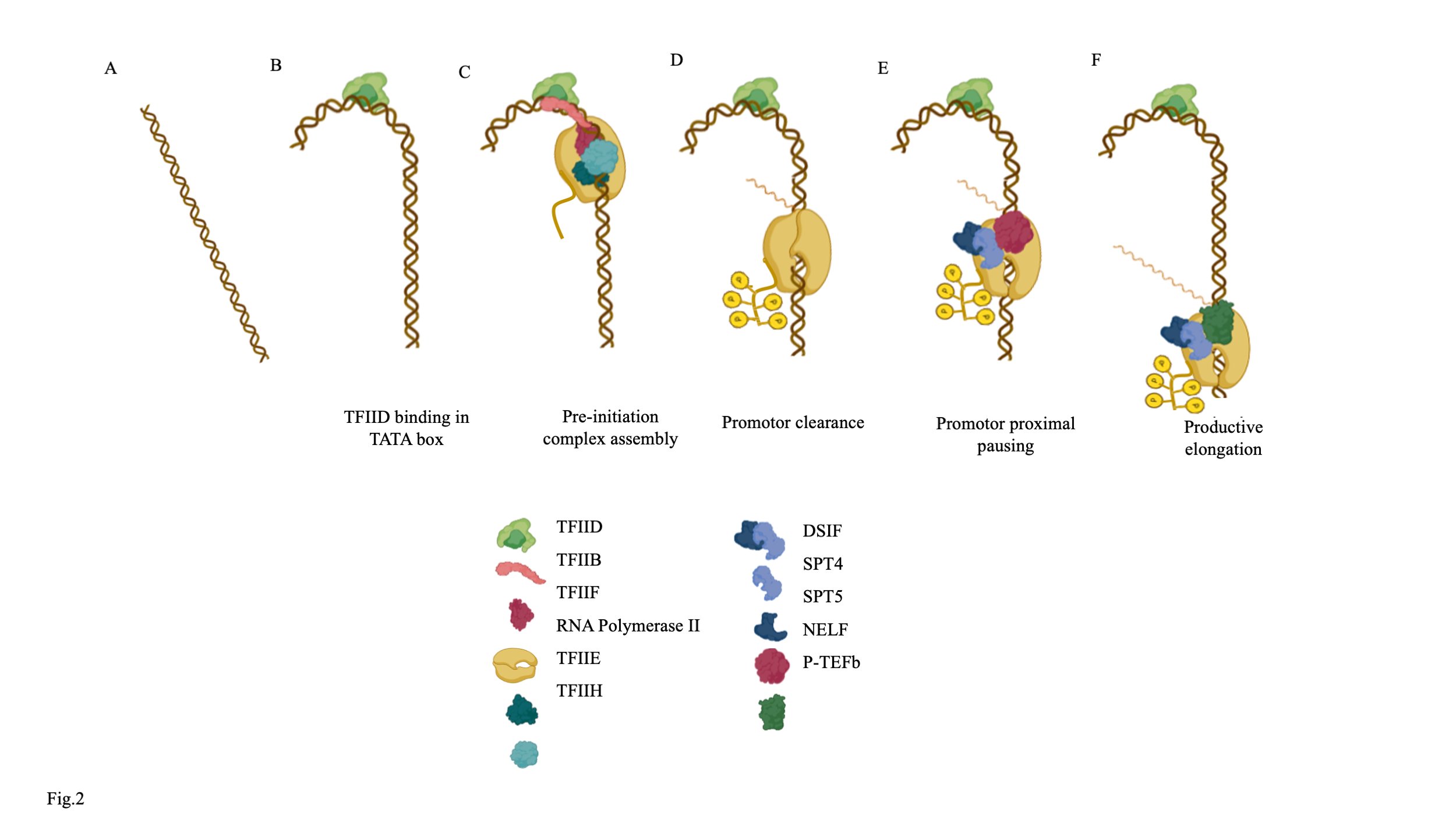
Fig. 2: Role of DRB sensitivity-inducing factor (DSIF) comprising SPT4 and SPT5 in the transcription machinery. (A) A section of a gene (B) TFIID binds to the TATA box, bends the DNA to begin recruiting the transcription machinery. (C) RNA polymerases II and the other general transcription factors, including TFIIB, TFIIF, TFIIE, and TFIIH, bind upstream of the transcription start site and form the pre-initiation complex. (D) Phosphorylation of the C-terminal domain (CTD) of RNA polymerase II leads to promoter clearance. (E) The interaction of DSIF complex with the negative elongation factor (NELF) forms the basis for promoter-proximal pause. (F) P-TEFb is a kinase that phosphorylates DSIF, NELF, and the CTD of Pol II, this reactivating the paused Pol II for a productive elongation.
SPT5 plays essential roles by interacting with various RNA processing and modification systems enroute to transcription [55]. It has been reported to resemble the Pol II CTD; the SPT5-CTD can also recruit the capping complex during transcription onto the newly formed RNA (Fig. 3A) [56]. In contrast to RNAP II CTD, in which phosphorylation of Ser 5 plays an essential role in the Guanylyl transferase (GTase) recruitment to the capping site, an unphosphorylated CTD of SPT5 is crucial to recruit the capping complex [57]. Upon phosphorylation at the Thr 1 position of SPT5-CTD, uncoupling of the capping complex from RNAP II occurs, resulting in release of the capping apparatus. Another independent study performed on yeast showed similar results and reported co-purification of capping enzymes Cet1 and Ceg1. Additionally, Abd1 with additional methyltransferase activity was co-purified along with the capping enzymes cet1 and ceg1(Fig. 3B) [58]. In addition to playing a role in 5’ processing activity, SPT5 is also critical in regulating the 3’ processing. Mayer et al., reported in their study on the importance of SPT5-CTR in the recruitment of pre-mRNA cleavage factor I (CFI) at the 3’end of S. cerevisiae [59]. In addition, the group also reported CFI in the SPT5-CTR pull-down in vitro. The mutant SPT5 Δ CTR showed a reduced occupancy of the elongation factor Paf1 and the CFI. NusG is reported to have a similar function in the Rho-dependent termination of transcription by the bacteria [60]. In addition to recruitment of various pre-mRNA processing intermediates onto RNA, SPT5 also plays a role in recruitment of complexes such as Paf1, involved in histone modifications, including methylation [61].
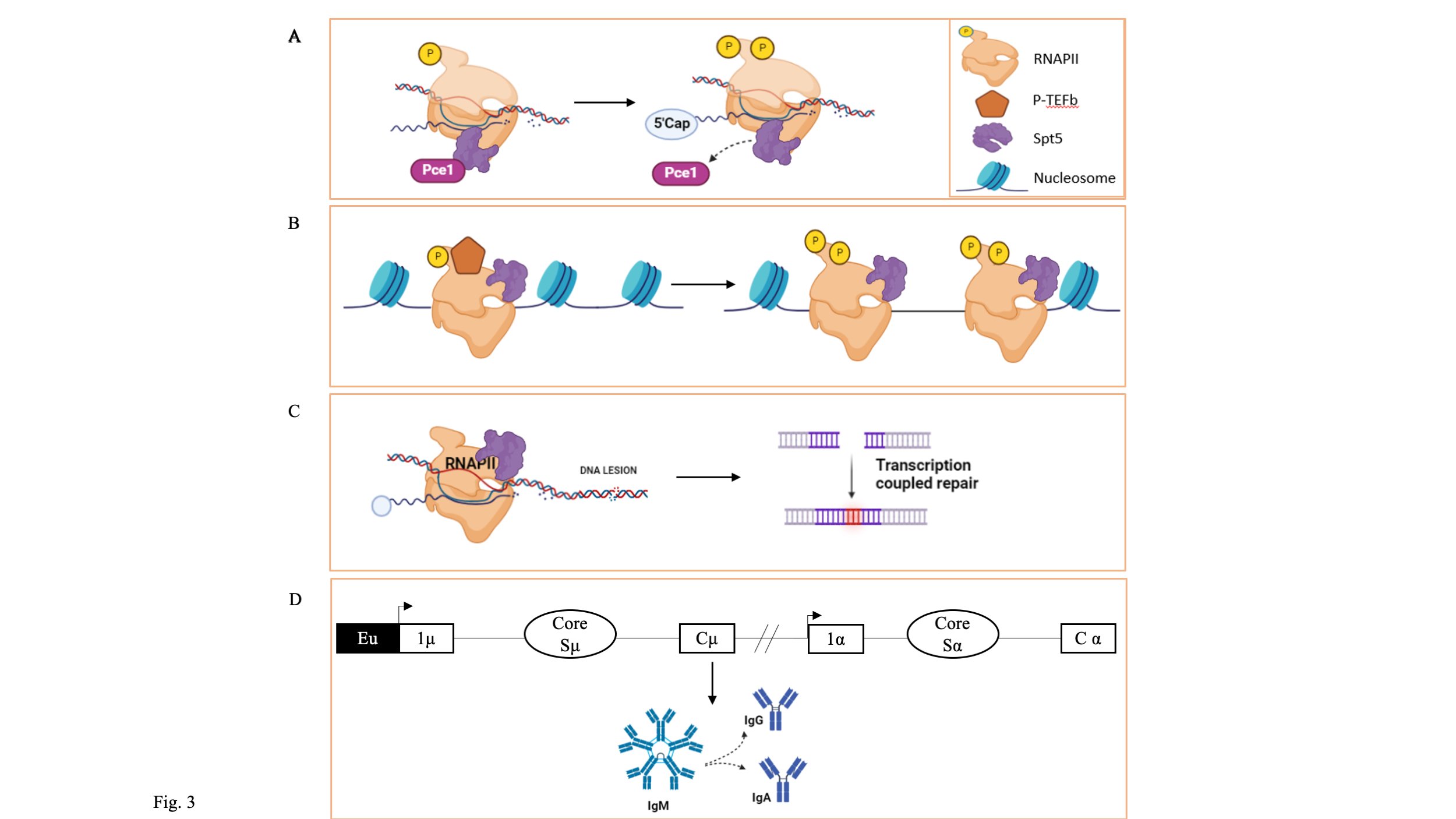
Fig. 3: Role of SPT5 in different transcription processes. (A) SPT5 serves as a platform for protein recruitment for components of the capping complex, such as Pce1, and assists them in attaching it to RNAPII to cap the newly produced RNA. (B)SPT5 promotes transcription by the nucleosome by acting as a nucleosome modifier. (C) SPT5 affects transcription at the location of DNA lesions, contributing to transcription-coupled DNA repair (TCR). (D) The immunoglobulin class switch recombination in B-cells is mediated by SPT5, a component of the DSIF complex.
Eukaryotic DNA is packed as a nucleosome, with the DNA tightly coiled around histones and the nucleosome acting as a transcription barrier [62]. SPT5 has been reported to facilitate transcription across the nucleosome barrier by acting as a histone chaperone similar to the function of FACT and SPT6 [63][64][65]. This function is attributed to the intrinsically disorganized region in the N-terminus of SPT5. The N-terminal is thought to initiate the development and binding of DNA to H2A and H2B, thus facilitating the transcription through the nucleosome (Fig 3B) [66].
In addition to serving as a recruitment platform for several RNA processing intermediates, SPT5 is also known to play a significant role in suppressing transcription-coupled DNA repair (TCR) [67]. TCR is one of the two pathways involved in nucleotide excision repair (NER), the other being global genome repair (GGR) [68]. It has been reported that the SPT5-CTR is essential in suppressing the Rad26-independent TCR. The yeast cell with a mutant SPT5-CTR showed an enhanced transcriptional activity compared to the wild-type in the yeast strain lacking Rad16 and Rad26 involved in the GGR and TCR, respectively [69]. Also, SPT4, part of the DSIF complex, showed a similar phenotype. Similar toSPT5-CTR, SPT4 also showed the ability to suppress the Rad26-independent CTR (Fig. 3C) [70].
Several other studies suggest the crucial role of SPT5 and its partner SPT4 in non-homologous end joining (NHEJ) and homologous recombination (HR), two major DNA repairing mechanisms. Inhibition of SPT5 resulted in a severely compromised NHEJ repair pathway of the damaged DNA in H1299d-Ad3. PCR analysis of the damaged DNA region revealed significant resurrection and lengthy insertion. A similar effect was seen in CH12F3-2A, which showed an increased sensitivity towards the ionizing radiation. A similar decline in the potential to repair DSBs was seen in GM7166VA cell lines, which further validates its role in TCR. These roles played by SPT5 in DNA repair mechanisms are essential for class switch recombination (CSR) in B cell, required for switching antibodies from IgM to IgA.[71][72]. CH12F3-2A upon induction with CIT (CD40L, IL4, and TGFβ) resulted in switching from IgM to IgA. Reduced antibody switching was the outcome of SPT5/SPT4 inhibition, which is associated with a decline in the S region cleavage following SPT5 inhibition (Fig. 3D) [73].
Further, SPT5 plays a critical role in the PTM of several histones and modifications of the nucleosome complex. SPT5 knock-down (KD) is known to downregulate the basal and induced level of H3K4me3 in the NFKB-regulated gene and downregulate the level of H4K5Ac at the promoter-proximal site (Fig. 4D)[74]. Downregulation in the H3K4me3 & H4K5Ac results in an increased promoter-proximal pausing, suggesting SPT5 is essential for acetylation and methylation of the nucleosome at the promoter-proximal site. These PTM regulated by the SPT5 are essential in recruiting TFIID and releasing TFIIE and the mediator complex from RNAP II [75]. In addition to controlling the methylation and acetylation of the histone markers, SPT5 is also methylated by a number of arginine methyltransferases, including PRMT1 and PRMT5, which affects its interaction with RNAP II and its potential for further elongation [76].
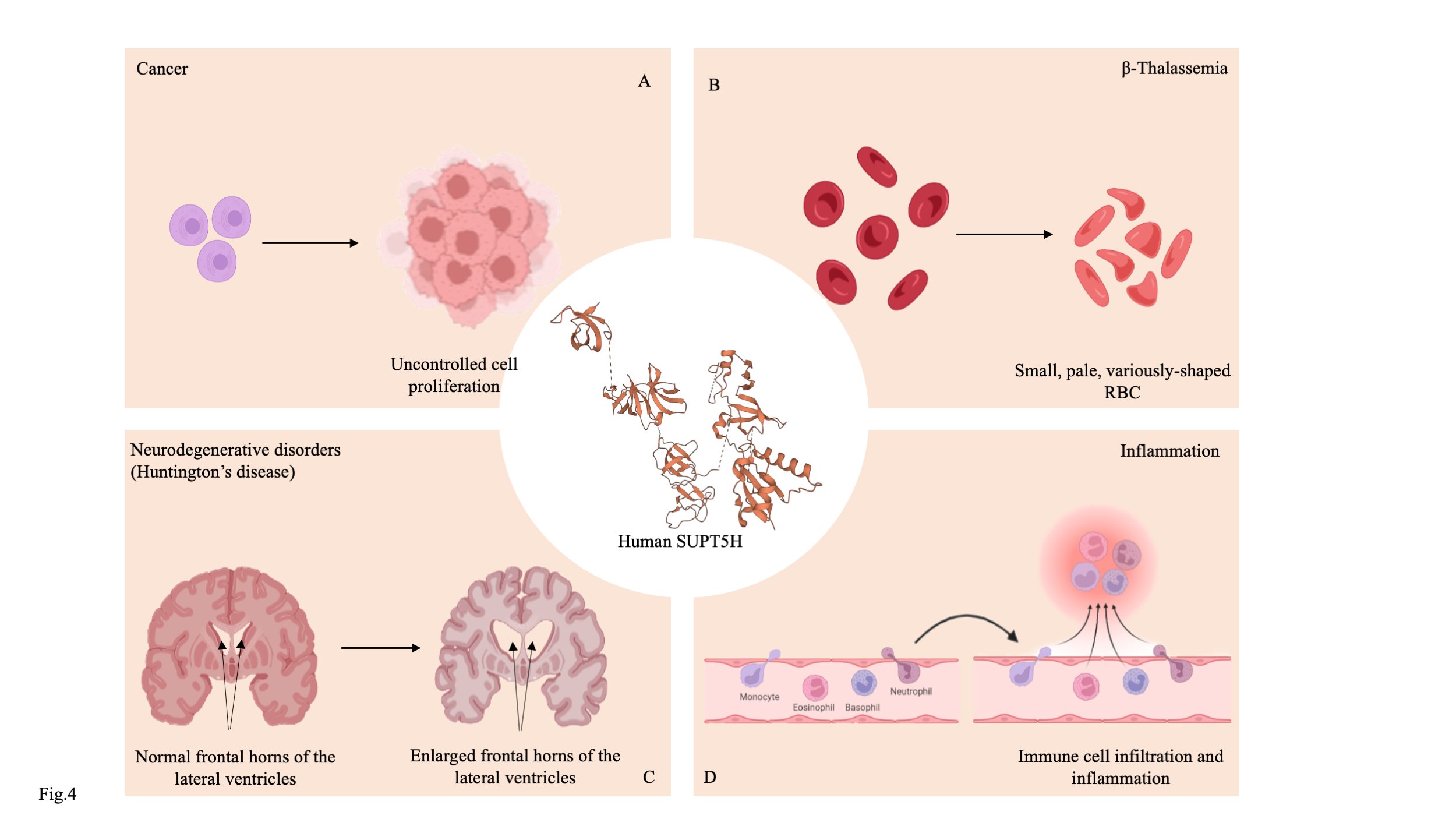
Fig. 4: Roles of SUPT5H/SPT5 in disease pathogenesis. (A) SPT5 supports the continuous cell proliferation of cancer cells by activating hTERT, an essential element for telomere maintenance. (B) Haploinsufficient mutation in the SUPT5H gene results in the downregulation of the HBB gene, causing β-thalassemia. (C) SPT4/SPT5 complex plays a role in synthesizing proteins with long stretches of CAG, producing insoluble proteins that cause neurodegenerative disorders, including Huntington's disease. (D) SPT5 plays a crucial role in the transcription of inflammation and stress-induced genes. SPT5 (PDB ID: 6GML).
SPT5 in non-coding RNA synthesis
Non-coding RNA refers to the large class of non-protein coding RNAs that are known to activate and regulate a variety of protein-coding genes and active genes. Since non-coding RNA is involved in a variety of cellular and developmental processes, it has attracted a lot of interest in recent years [77]. Many non-coding RNAs are known to be involved in multiple disease pathogenesis, while others are engaged in the regulation of diseases. Therefore, it is very interesting to study the biology of non-coding RNA.
The SPT5-SPT4 complex is also reported to regulate the expression level of non-coding RNA in a manner similar to how it controls the synthesis of protein-coding RNA. The role of SPT4 and SPT5 in controlling the expression of the germline genome was demonstrated by a study by Owsain et al. in the Paramecium tetraurelia. SPT4-SPT5 complex played a significant role in the genome rearrangement, necessary for the development of P.tetraureliaI, by being required for the synthesis of scnRNA that is required for generating sRNA [78]. Another group in an independent research revealed that SPT5 suppression reduced the expression of eRNA in both Drosophila and humans [79][72]. The non-coding RNA known as enhancers is crucial for several regulatory processes connected to the activation of genes [80]. SPT5 plays a critical role here by providing much-needed stability to the RNAP II at the promoter region of enhancers. Due to decreased stability, RNAPII is significantly more susceptible to premature termination at the enhancer location of the promoter than it is at the site of the protein-coding gene. In addition to SPT5 directly controlling eRNA synthesis, it is also known that eRNA indirectly controls SPT5 activity by downregulating or upregulating various of inhibitors and activators. eRNAs play an important role in releasing NELF, resulting in a transition elongation complex [81]. SPT5 is crucial for chromatin accessibility, as was described in the preceding section [82]. Inhibition of SPT5 restricted access to enhancers and lowered SWI/SNF occupancy, both of which are necessary for chromatin remodelling [83].
SPT5 Role in Disease Pathogenesi
The human analog of SPT5, SUPT5H has been linked to a number of diseases. According to Chen et al., SUPT5H acts as an activator for hTERT and increases its reverse transcriptase activity [84]. Telomere maintenance in cancer cells depends on increased hTERT activity [85]. The proliferative and metastatic capacity of colon cancer cells was reduced by SPT5 suppression by shRNA. A similar study by Bilal et al. reported the role of SUPT5H in breast cancer proliferation. The metastatic potential was reduced and the survival-related MAPK signalling was significantly downregulated when SUPT5H was inhibited [86].Additionally, SUPT5H was interacting with the Paf complex, which is known to be involved in the development of tumours. According to a recent study, SUPT5H plays a prognostic function in predicting the likelihood of developing intrahepatic cholangiocarcinoma (ICC) (Fig. 4A) [87].
SPT5 not only plays essential role in cancer progression, but also plays a crucial function in HIV transcription. It interacts with HIV-Tat1 and preserves its stability at the transcription site. SPT5 knockdown lowered pol II and Cdk9 occupancy and accelerated HIV entry into latency [88][89]. Additionally, it is known that SUPT5H controls the HBB gene, which codes for the beta-globin gene. The HBB protein, which is accountable for β-thalassemia, was downregulated as a result of a haploinsuffficient mutation on the SUPT5H (Fig. 4B, D) [90].
Recent research reports the SPT4-SPT5 complex's involvement in neurodegenerative diseases. They have reportedly contributed to the production of insoluble proteins that cause neurodegenerative diseases by synthesising proteins with lengthy CAG stretches [91]. Frontotemporal dementia (FTD), Huntington's disease (HD), and amyotrophic lateral sclerosis (ALS) have all been linked to the SPT4/SPT5 complex [92]. RNA interference-mediated inhibition of Spt4/Spt5 reduced the amount of mutant Htt protein having extensive CAG stretches The short CAG protein, however, is unaffected by the aforementioned interference( Fig. 4C)[93].
Discussion
SPT5 performs important functions in the transcription process from the promoter-proximal pause to the termination, making it clear that SPT5 is a crucial part of the transcription machinery in all three domains of life. It also associates with different RNA modifiers, causing varying RNA modifications during the course of transcription. However, there are still a number of unanswered concerns regarding the precise function of SPT5 at various stages of disease aetiology and regulation. The global and gene-specific roles of SPT5 in transcription regulation is the most important issues that require further study. It has been discovered that blocking SPT5 in yeast reduced the expression of only few sets of essential genes that are important for its development. However, blocking SPT5 in human and Drosophila caused to have reduced expression of majority of their genes [94].. Therefore, studies focusing on SPT5's overall and gene-specific roles in transcriptional control are required.
It is well known that SPT5 is essential for controlling the both RNAP I, which is in charge of rRNA synthesis, and RNAP II[95][96]. Although it is well known that SPT5 affects RNAP I in both a positive and a negative manner, further research is required to define the molecular mechanism and the protein interactome [97].
Additionally, more research needs to be done on the SPT5 phosphorylation landscape. Therefore, it is necessary to look into how phosphorylation at various sites affect the transcriptional machinery in order to better our understanding. The role of SPT5 in various metabolic stressors, how it responds to them, and the crucial factors that cause it to switch from a regulator to an activator at the promoter-proximal site are all things that need to be understood in order for this process to proceed. Numerous studies have emphasised the crucial function of SPT5 and the DSIF complex in controlling non-coding RNA involved in a variety of processes, such as development, gene regulation, and the activation of the target gene via enhancer function, in addition to controlling transcription at the protein-coding genes. Some studies have highlighted the role of SPT5 in the epigenetic landscape, as SPT5 inhibition resulted in altered expression of acetylation and the methylation markers. Therefore, thorough research showing the precise part SPT5 plays in altering the epigenetic landscape is required.
Several studies highlighted the oncogenic role of SUPT5H in the activation of MAPK, Myc, and hTERT proteins [84][98] [99]. Apoptosis, cell cycle arrest, and senescence are just a few of the pathways that are activated when SUPT5H is knocked down that are important in preventing cell proliferation [84] [99][100]. Different SUPT5H knocked-down cell lines showed differential outcomes regarding apoptosis or senescence upon SUPT5H inhibition. Therefore, it is necessary to investigate the cause of either one or both pathways becoming engaged. SPT5 is also involved in the pathogenesis of a number of other illnesses, including inherited illnesses like thalassemia and neurological diseases. However, research aimed at novel paradigms in this direction is still needed to fully understand its role as a transcriptional regulator in these disorders.
Acknowledgements
We gratefully acknowledge South Asian University, Rajpur Road, Maidan Garhi, New Delhi, India, for the institutional support, Vivek Pandey would like to acknowledge the Department of Biotechnology, INDIA, for providing Senior Research Fellowship to accomplish the study.
Author Contributions
VP and YRP conceptualized the review, VP did the review writing, VP, SP & YRP performed the editing, VP and SP made figures.
Funding Sources
We thank the Department of Biotechnology, Government of India & South Asian University.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Cramer P, Bushnell DA, Kornberg RD: Structural basis of transcription RNA polymerase II at 2.8 angstrom resolution. Science 2001; 292: 1863-1876.
https://doi.org/10.1126/science.1059493 |
| 2 | Cramer P: Organization and regulation of gene transcription. Nat 2019; 573: 45-54.
https://doi.org/10.1038/s41586-019-1517-4 |
| 3 | Core L, Adelman K: Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 2019; 960-982.
https://doi.org/10.1101/gad.325142.119 |
| 4 | Chen FX, Smith ER, Shilatifard A: Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2018; 19: 464-478.
https://doi.org/10.1038/s41580-018-0010-5 |
| 5 | Hantsche M, Cramer P: The Structural Basis of Transcription: 10 Years After the Nobel Prize in Chemistry. Angew Chemie Int Ed 2016; 55: 15972-15981.
https://doi.org/10.1002/anie.201608066 |
| 6 | Hartzog GA, Fu J: The Spt4-Spt5 complex: A multi-faceted regulator of transcription elongation. Biochim Biophys Acta-Gene Regul Mech 2013; 1829: 105-115.
https://doi.org/10.1016/j.bbagrm.2012.08.007 |
| 7 | Mooney RA, Schweimer K, Rösch P, Gottesman M, Landick R: Two Structurally Independent Domains of E.coli NusG Create Regulatory Plasticity via Distinct Interactions with RNA Polymerase and Regulators. J Mol Biol 2009; 391: 341-358.
https://doi.org/10.1016/j.jmb.2009.05.078 |
| 8 | Hartzog GA, Wada T, Handa H, Winston F: Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 1998; 12: 357-369.
https://doi.org/10.1101/gad.12.3.357 |
| 9 | Swanson MS, Winston F: SPT4, SPT5 and SPT6 interactions: Effects on transcription and viability in Saccharomyces cerevisiae. Genetics 1992; 132: 325-336.
https://doi.org/10.1093/genetics/132.2.325 |
| 10 | Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose H, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handaet H: DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 1998; 12: 343-356.
https://doi.org/10.1101/gad.12.3.343 |
| 11 | Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H: Structure and function of the human transcription elongation factor DSIF. J Biol Chem 1999; 274: 8085-8092.
https://doi.org/10.1074/jbc.274.12.8085 |
| 12 | Winston F, Chaleff DT, Valent B, Fink GR: Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 1984; 107: 179-197.
https://doi.org/10.1093/genetics/107.2.179 |
| 13 | Fassler JS, Winston F: Isolation and Analysis of a Novel Class of Suppressor of Ty Insertion Mutations in Saccharomyces Cerevisiae. Genetics 1988; 118: 203. Accessed April 29, 2018.
https://doi.org/10.1093/genetics/118.2.203 |
| 14 | Malone EA, Fassler J, Winston F: Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Published online 2008.
|
| 15 | Swanson MS, Malone EA, Winston F: SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 1991; 11: 3009-3019.
https://doi.org/10.1128/MCB.11.6.3009 |
| 16 | Dollinger R, Gilmour DS: Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans. J Mol Biol 2021; 433: 166897.
https://doi.org/10.1016/j.jmb.2021.166897 |
| 17 | Decker TM: Mechanisms of Transcription Elongation Factor DSIF (Spt4-Spt5). J Mol Biol 2020; 433: 166657-166657.
https://doi.org/10.1016/j.jmb.2020.09.016 |
| 18 | Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter T, Brannan K, Bentley DL: Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a "Sitting Duck Torpedo" Mechanism. Mol Cell 2019; 76: 896-908
https://doi.org/10.1016/j.molcel.2019.09.031 |
| 19 | Ivanov D, Kwak YT, Guo J, Gaynor RB: Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 2000; 20: 2970-2983.
https://doi.org/10.1128/MCB.20.9.2970-2983.2000 |
| 20 | Komori T, Inukai N, Yamada T, Yamaguchi Y, Handa H: Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes to Cells 2009; 14: 343-354.
https://doi.org/10.1111/j.1365-2443.2008.01273.x |
| 21 | Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi , Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi M A, Sandelin A, Chalk A M, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C: Antisense transcription in the mammalian transcriptome. Science 2005; 309: 1564-1566.
https://doi.org/10.1126/science.1112009 |
| 22 | Park K, Zhong J, Jang JS, Kim J, Kim H, Lee J, Kim J: ZWC complex-mediated SPT5 phosphorylation suppresses divergent antisense RNA transcription at active gene promoters. Nucleic Acids Res 2022; 50: 3835-3851.
https://doi.org/10.1093/nar/gkac193 |
| 23 | Werner F: A Nexus for Gene Expression-Molecular Mechanisms of Spt5 and NusG in the Three Domains of Life. J Mol Biol 2012; 417: 13-27.
https://doi.org/10.1016/j.jmb.2012.01.031 |
| 24 | Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P: Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J 2011; 30: 1302-1310.
https://doi.org/10.1038/emboj.2011.64 |
| 25 | Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H: Structure and function of the human transcription elongation factor DSIF. J Biol Chem 1999; 274: 8085-8092.
https://doi.org/10.1074/jbc.274.12.8085 |
| 26 | Hirtreiter A, Damsma GE, Cheung ACM, Klose D, Grohmann D, Vojnic E, Martin ACR, Cramer P, Werner F: Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res 2010; 38: 4040-4051.
https://doi.org/10.1093/nar/gkq135 |
| 27 | Kyrpides NC, Woese CR, Ouzounis CA: KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci 1996; 21: 425-426.
https://doi.org/10.1016/S0968-0004(96)30036-4 |
| 28 | Steiner T, Kaiser JT, Marinkoviç S, Huber R, Wahl MC: Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J 2002; 21: 4641-4653.
https://doi.org/10.1093/emboj/cdf455 |
| 29 | Reay P, Yamasaki K, Terada T, Kuramitsu S, Shirouzu M, Yokoyama S: Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins 2004; 56: 40-51.
https://doi.org/10.1002/prot.20054 |
| 30 | Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS: RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A 2011; 108: 546-550.
https://doi.org/10.1073/pnas.1013828108 |
| 31 | Meyer PA, Li S, Zhang M, Yamada K, Takagi Y, Hartzog GA, Fu J: Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5 Mol Cell Biol 2015; 35: 3354-3369.
https://doi.org/10.1128/MCB.00520-15 |
| 32 | Lasko P: Tudor domain. Curr Biol 2010; 20: 666-667.
https://doi.org/10.1016/j.cub.2010.05.056 |
| 33 | Li W, Giles C, Li S: Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res 2014; 42: 7069-7083.
https://doi.org/10.1093/nar/gku333 |
| 34 | Hu S, Peng L, Xu C, Wang Z, Song A, Chen FX: SPT5 stabilizes RNA polymerase II, orchestrates transcription cycles, and maintains the enhancer landscape. Mol Cell 2021; 81: 4425-4439.
https://doi.org/10.1016/j.molcel.2021.08.029 |
| 35 | Bernecky C, Plitzko JM, Cramer P: Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol 2017; 24: 809-815.
https://doi.org/10.1038/nsmb.3465 |
| 36 | Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, Handa H, Ish-Horowicz D: Locus-Specific Requirements for Spt5 in Transcriptional Activation and Repression in Drosophila Results and Discussion Identification of a Novel Drosophila Mutation with a Maternal Segmentation Defect To discover novel maternal mutations affecting embry. Curr Biol 2004; 14: 1680-1684 .
https://doi.org/10.1016/j.cub.2004.08.066 |
| 37 | Chen H, Contreras X, Yamaguchi Y, Handa H, Peterlin BM, Guo S: Repression of RNA Polymerase II Elongation In Vivo Is Critically Dependent on the C-Terminus of Spt5. PLoS One 2009; 4: 1-11.
https://doi.org/10.1371/journal.pone.0006918 |
| 38 | Swanson MS, Malone EA, Winston F: SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 1991; 11: 3009-3019.
https://doi.org/10.1128/MCB.11.6.3009 |
| 39 | Diamant G, Bahat A, Dikstein R: The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes. Nat Commun 2016; 7: 1-13.
https://doi.org/10.1038/ncomms11547 |
| 40 | Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H: P-TEFb-Mediated Phosphorylation of hSpt5 C-Terminal Repeats Is Critical for Processive Transcription Elongation. Mol Cell 2006; 21: 227-237.
https://doi.org/10.1016/j.molcel.2005.11.024 |
| 41 | Shetty A, Kallgren SP, Demel C, Maier KC, Spatt D, Alver BH Cramer P, Park PJ, Winston F: Spt5 Plays Vital Roles in the Control of Sense and Antisense Transcription Elongation. Mol Cell 2017; 66: 77-88.
https://doi.org/10.1016/j.molcel.2017.02.023 |
| 42 | Mueller CL, Porter SE, Hoffman MG, Jaehning JA: The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 2004;14: 447-456.
https://doi.org/10.1016/S1097-2765(04)00257-6 |
| 43 | Qiu Y, Gilmour DS: Identification of regions in the Spt5 subunit of DRB sensitivity-inducing factor (DSIF) that are involved in promoter-proximal pausing. J Biol Chem 2017; 292: 5555-5570.
https://doi.org/10.1074/jbc.M116.760751 |
| 44 | Bernecky C, Plitzko JM, Cramer P: Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol 2017; 24: 809-815.
https://doi.org/10.1038/nsmb.3465 |
| 45 | Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, Parrinello H, Cuvier O, Benkirane M: Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 2014; 5: 5531-5541.
https://doi.org/10.1038/ncomms6531 |
| 46 | Vos SM, Farnung L, Urlaub H, Cramer P: Structure of paused transcription complex Pol II-DSIF-NELF. N 2010; 107: 11301-11306.
https://doi.org/10.1073/pnas.1000681107 |
| 47 | Missra A, Gilmour DS: Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Nature 2018; 560: 607-612.
|
| 48 | Fong N, Sheridan RM, Ramachandran S, Bentley DL: The pausing zone and control of RNA polymerase II elongation by Spt5: Implications for the pause-release model. Mol Cell 2022; 82: 3632-3645 .
https://doi.org/10.1016/j.molcel.2022.09.001 |
| 49 | Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN: The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J 2008; 27: 1907-1918.
https://doi.org/10.1038/emboj.2008.121 |
| 50 | Sansó M, Levin RS, Lipp JJ, Wang LVY, Greifenberg AK, Quezada EM, Ali A, Ghosh A, Larochelle S, Rana TM, Geyer M, Tong L, Shokat KM, Fisher RP: P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev 2016; 30: 117-131.
https://doi.org/10.1101/gad.269589.115 |
| 51 | Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P: Structure of activated transcription complex Pol II-DSIF-PAF-SPT6 Nature 2018; 560: 607-612.
https://doi.org/10.1038/s41586-018-0440-4 |
| 52 | Aoi Y, Takahashi Y hei, Shah AP, Iwanaszko M, Rendleman EJ, Khan NH, Cho B, Goo YA, Ganesan S, Kelleher NL, Shilatifard A: SPT5 stabilization of promoter-proximal RNA polymerase II. Mol Cell 2021; 81: 4413-4424.
https://doi.org/10.1016/j.molcel.2021.08.006 |
| 53 | Shetty A, Kallgren SP, Demel C, Maier KC, Spatt D, Alver BH, Cramer P, Park PJ, Winston F: Spt5 Plays Vital Roles in the Control of Sense and Antisense Transcription Elongation. Mol Cell 2017; 66: 77-88
https://doi.org/10.1016/j.molcel.2017.02.023 |
| 54 | Pavri R, Gazumyan A, Jankovic M, Virgilio MD, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, San-Martin AR, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC: Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5 Cell 2010; 143: 122-133.
https://doi.org/10.1016/j.cell.2010.09.017 |
| 55 | Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, Hartzog GA: Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 2003; 23: 1368-1378.
https://doi.org/10.1128/MCB.23.4.1368-1378.2003 |
| 56 | Wen Y, Shatkin AJ: Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 1999; 13: 1774.
https://doi.org/10.1101/gad.13.14.1774 |
| 57 | Chu C, Das K, Tyminski JR, Bauman JD, Guan R, Qiu W, Montelione GT, Arnold E, Shatkin AJ: Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc Natl Acad Sci USA 2011; 108: 10104-10108.
https://doi.org/10.1073/pnas.1106610108 |
| 58 | Doamekpor SK, Sanchez AM, Schwer B, Shuman S, Lima CD: How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev 2014; 28: 1323-1336.
https://doi.org/10.1101/gad.242768.114 |
| 59 | Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P: The spt5 C-terminal region recruits yeast 3' RNA cleavage factor I. Mol Cell Biol 2012; 32: 1321-1331.
https://doi.org/10.1128/MCB.06310-11 |
| 60 | Valabhoju V, Agrawal S, Sen R: Molecular Basis of NusG-mediated Regulation of Rho-dependent Transcription Termination in Bacteria. J Biol Chem 2016; 291: 22386-22403.
https://doi.org/10.1074/jbc.M116.745364 |
| 61 | Lu C, Tian Y, Wang S, Su Y, Mao T, Huang T, Chen Q, Xu Z, Ding Y: Phosphorylation of SPT5 by CDKD;2 Is Required for VIP5 Recruitment and Normal Flowering in Arabidopsis thaliana. Plant Cell 2017; 29: 277-291.
https://doi.org/10.1105/tpc.16.00568 |
| 62 | McGinty RK, Tan S: Nucleosome structure and function. Chem Rev 2015; 115: 2255-2273.
https://doi.org/10.1021/cr500373h |
| 63 | Jeronimo C, Poitras C, Robert F: Histone Recycling by FACT and Spt6 during Transcription Prevents the Scrambling of Histone Modifications. Cell Rep 2019; 28: 1206-1218.
https://doi.org/10.1016/j.celrep.2019.06.097 |
| 64 | Bobkov GOM, Huang A, van den Berg SJW, Mitra S, Anselm E, Lazou V, Schunter S, Feederle R, Imhof A, Lusser A, Jansen LET, Patrick: Spt6 is a maintenance factor for centromeric CENP-A. Nat Commun 2020; 11: 1-14.
https://doi.org/10.1038/s41467-020-16695-7 |
| 65 | Duina AA: Histone Chaperones Spt6 and FACT: Similarities and Differences in Modes of Action at Transcribed Genes. Genet Res Int 2011: 1-12.
https://doi.org/10.4061/2011/625210 |
| 66 | Evrin C, Serra-Cardona A, Duan S, Mukherjee PP, Zhang Z, Labib KPM: Spt5 histone binding activity preserves chromatin during transcription by RNA polymerase II. EMBO J 2022; 41: 109783 .
https://doi.org/10.15252/embj.2021109783 |
| 67 | Ding B, LeJeune D, Li S: The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem 2010; 285: 5317-5326.
https://doi.org/10.1074/jbc.M109.082818 |
| 68 | Spivak G: Transcription-coupled repair: an update. Arch Toxicol 2016; 90: 2583-2594.
https://doi.org/10.1007/s00204-016-1820-x |
| 69 | Li S, Ding B, LeJeune D, Ruggiero C, Chen X, Smerdon MJ: The roles of Rad16 and Rad26 in repairing repressed and actively transcribed genes in yeast. DNA Repair (Amst) 2007; 6: 1596-1606.
https://doi.org/10.1016/j.dnarep.2007.05.005 |
| 70 | Jansen LET, Den Dulk H, Brouns RM, De Ruijter M, Brandsma JA, Brouwer J: Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J 2000;19(23):6498-6507.
https://doi.org/10.1093/emboj/19.23.6498 |
| 71 | Stanlie A, Begum NA, Akiyama H, Honjo T: The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination. PLOS Genet 2012; 8: 1002675.
https://doi.org/10.1371/journal.pgen.1002675 |
| 72 | Fitz J, Neumann T, Steininger M, Wiedemann E, Garcia AC, Athanasiadis A, Schoeberl UE, Pavri R: Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nat Genet 2020; 52: 505-515.
https://doi.org/10.1038/s41588-020-0605-6 |
| 73 | Kracker S, Radbruch A: Immunoglobulin class switching: in vitro induction and analysis. Methods Mol Biol 2004; 271: 149-159.
https://doi.org/10.1385/1-59259-796-3:149 |
| 74 | Diamant G, Bahat A, Dikstein R: The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes. Nat Commun 2016; 7: 1-13.
https://doi.org/10.1038/ncomms11547 |
| 75 | Compe E, Genes CM, Braun C, Coin F, Egly JM: TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription. Nat Commun 2019; 10: 2084-2098.
https://doi.org/10.1038/s41467-019-10131-1 |
| 76 | Kwak YT, Guo J, Prajapati S, Park K, Surabhi RM, Miller B, Gehrig P, Gaynor RB: Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell 2003; 11: 1055-1066.
https://doi.org/10.1016/S1097-2765(03)00101-1 |
| 77 | Mattick JS, Makunin IV: Non-coding RNA. Hum Mol Genet 2006; 15 Spec No 1: 172-179.
https://doi.org/10.1093/hmg/ddl046 |
| 78 | Gruchota J, Wilkes CD, Arnaiz O, Sperling L, Nowak JK: A meiosis-specific Spt5 homolog involved in non-coding transcription. Nucleic Acids Res 2017; 45: 4722-4732.
https://doi.org/10.1093/nar/gkw1318 |
| 79 | Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, Adelman K: Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 2018; 32: 26-41.
https://doi.org/10.1101/gad.309351.117 |
| 80 | Rahnamoun H, Orozco P, Lauberth SM: The role of enhancer RNAs in epigenetic regulation of gene expression. Transcription 2020; 11: 19-25.
https://doi.org/10.1080/21541264.2019.1698934 |
| 81 | Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK: Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 2014; 56: 29-42.
https://doi.org/10.1016/j.molcel.2014.08.023 |
| 82 | Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010; 107: 21931-21936
https://doi.org/10.1073/pnas.1016071107 |
| 83 | Iurlaro M, Stadler MB, Masoni F, Jagani Z, Galli GG, Schübeler D: Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet 2021; 53: 279-287.
https://doi.org/10.1038/s41588-020-00768-w |
| 84 | Chen R, Zhu J, Dong Y, He C, Hu X: Suppressor of Ty homolog-5, a novel tumor-specific human telomerase reverse transcriptase promoter-binding protein and activator in colon cancer cells. Oncotarget 2015; 6: 32841-32855.
https://doi.org/10.18632/oncotarget.5301 |
| 85 | Hannen R, Bartsch JW: Essential roles of telomerase reverse transcriptase hTERT in cancer stemness and metastasis. FEBS Lett 2018; 592: 2023-2031.
https://doi.org/10.1002/1873-3468.13084 |
| 86 | Lone BA, Ahmad F, Karna SKL, Pokharel YR: SUPT5H Post-Transcriptional Silencing Modulates PIN1 Expression, Inhibits Tumorigenicity, and Induces Apoptosis of Human Breast Cancer Cells. Cell Physiol Biochem 2020; 54: 928-946.
https://doi.org/10.33594/000000279 |
| 87 | Zou W, Wang Z, Zhang X, Xu S, Wang F, Li , Deng Z, Wang J, Pan K, Ge X, Li C, Liu R, Hu M: PIWIL4 and SUPT5H combine to predict prognosis and immune landscape in intrahepatic cholangiocarcinoma. Cancer Cell Int 2021; 21: 1-14.
https://doi.org/10.1186/s12935-021-02310-2 |
| 88 | Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J: Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol 2002; 22: 1079-1093.
https://doi.org/10.1128/MCB.22.4.1079-1093.2002 |
| 89 | Krasnopolsky S, Novikov A, Kuzmina A, Taube R: CRISPRi-mediated depletion of Spt4 and Spt5 reveals a role for DSIF in the control of HIV latency. Biochim Biophys Acta-Gene Regul Mech 2021; 1864: 194656.
https://doi.org/10.1016/j.bbagrm.2020.194656 |
| 90 | Achour A, Koopmann T, Castel R, Santen GWE, Hollander ND, Knijnenburg J, Ruivenkamp CAL, Arkesteijn SGK, Huurne JT, Bisoen S, Verschuren M, Vijfhuizen L, Schaap R, Grimbergen A, Slomp J, Traeger-Synodinos J, Vrettou C, Pissard S, Galacteros F, Baas F, Harteveld CL: A new gene associated with a β-thalassemia phenotype: the observation of variants in SUPT5H. Blood 2020; 136: 1789-1793.
https://doi.org/10.1182/blood.2020005934 |
| 91 | Furuta N, Tsukagoshi S, Hirayanagi K, Ikeda Y: Suppression of the yeast elongation factor Spt4 ortholog reduces expanded SCA36 GGCCUG repeat aggregation and cytotoxicity. Brain Res 2019; 1711: 29-40.
https://doi.org/10.1016/j.brainres.2018.12.045 |
| 92 | Stoyas CA, La Spada AR: The CAG-polyglutamine repeat diseases: a clinical, molecular, genetic, and pathophysiologic nosology. Handb Clin Neurol 2018; 147: 143-170.
https://doi.org/10.1016/B978-0-444-63233-3.00011-7 |
| 93 | Bahat A, Lahav O, Plotnikov A, Leshkowitz D, Dikstein R:Targeting Spt5-Pol II by Small-Molecule Inhibitors Uncouples Distinct Activities and Reveals Additional Regulatory Roles. Mol Cell 2019; 76: 617-631.
https://doi.org/10.1016/j.molcel.2019.08.024 |
| 94 | Krishnan K, Salomonis N, Guo S: Identification of Spt5 target genes in zebrafish development reveals its dual activity in vivo. PLoS One 2008; 3: 1-13.
https://doi.org/10.1371/journal.pone.0003621 |
| 95 | Viktorovskaya O V, Appling FD, Schneider DA: Yeast Transcription Elongation Factor Spt5 Associates with RNA Polymerase I and RNA Polymerase II Directly. J Biol Chem 2011; 286: 18825-18833.
https://doi.org/10.1074/jbc.M110.202119 |
| 96 | Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M: RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci 2006; 103: 12707-12712.
https://doi.org/10.1073/pnas.0605686103 |
| 97 | Anderson SJ, Sikes ML, Zhang Y, French AH, Salgia S, Beyer AL, Nomura M, Schneider DA: The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J Biol Chem 2011; 286: 18816-18824.
https://doi.org/10.1074/jbc.M110.202101 |
| 98 | Baluapuri A, Hofstetter J, Stankovic DN, Endres T, Bhandare P, Vos SM, Adhikari B, Schwarz JD, Narain A, Vogt M, Wang S, Düster R, Jung LA, Vanselow JT, Wiegering A, Geyer M, Maric HM, Gallant P, Walz S, Schlosser A, Cramer P, Eilers M, Wolf E: MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol Cell 2019; 74: 674-687.
https://doi.org/10.1016/j.molcel.2019.02.031 |
| 99 | Lone BA, Ahmad F, Karna SKL, Pokharel YR: SUPT5H post-transcriptional silencing modulates PIN1 expression, inhibits tumorigenicity, and induces apoptosis of human breast cancer cells. Cell Physiol Biochem 2020; 54: 928-946.
https://doi.org/10.33594/000000279 |
| 100 | Komori T, Inukai N, Yamada T, Yamaguchi Y, Handa H: Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes to Cells 2009; 14: 343-354.
https://doi.org/10.1111/j.1365-2443.2008.01273.x |