Evaluation of SK-N-SH Cells as a Model for NMDA Receptor Induced Toxicity
bUniversity of Münster, GRK 2515, Chemical biology of ion channels (Chembion), Münster, Germany,
cCentre of Reproductive Medicine and Andrology, University of Münster, Domagkstraße 11, Münster, Germany,
dUniversity of Münster, Institute for Pharmaceutical and Medicinal Chemistry, Corrensstr. 48, Münster, Germany
Keywords
Abstract
Background/Aims:
Over the years, the number of patients with neurodegenerative diseases is constantly rising illustrating the need for new neuroprotective drugs. A promising treatment approach is the reduction of excitotoxicity induced by rising (S)-glutamate levels and subsequent NMDA receptor overactivation. To facilitate the search for new NMDA receptor inhibitors neuronal cell models are needed. In this study, we evaluated the suitability of human SK-N-SH cells to serve as a cell model for neurodegeneration induced by NMDA receptor overstimulation.Methods:
The cytoprotective effect of the unselective NMDA receptor blocker ketamine as well as the GluN2B-selective inhibitor WMS14-10 was evaluated utilizing different cell viability assays, such as endpoint (LDH, CCK-8, DAPI/FACS) and time dependent methods (bioimpedance).Results:
Non-differentiated as well as differentiated SK-N-SH cells express GluN1 and GluN2B subunits. Furthermore, 50 mM (S)-glutamate led to an instantaneous decrease in cell survival. Only application of unselective channel blocker ketamine could protect differentiated cells against this effect, while the selective inhibitor WMS14-10 did not significantly increase cell survival.Conclusion:
SK-N-SH cells show an increased sensitivity to (S)-glutamate mediated cytotoxicity with higher differentiation level, that is only partially induced by NMDA receptor overstimulation. Furthermore, we showed that only unselective NMDA receptor inhibition can partially reverse (S)-glutamate-induced toxicity.Introduction
Neurodegeneration is a common mechanism in Alzheimer’s (AD), Huntington’s or Parkinsons disease, which have an increasing prevalence as life expectancy rises [1–3]. One pathway of neurodegeneration is the process of excitotoxicity that is induced by uncontrolled release of (S)-glutamate from degenerated cells [4–7]. As a consequence, high concentrations of (S)-glutamate lead to overstimulation of glutamatergic receptors including the Ca2+ conducting N-methyl-D-aspartate (NMDA) receptors creating a self-enhancing apoptosis process due to Ca2+ overload [5, 8–10]. NMDA receptors are heterotetrameric ion channels, which are formed by two mandatory GluN1 subunits and different combinations of two GluN2 (GluN2A-D) and / or GluN3 (GluN3A-B) subunits [11, 12]. The composition of NMDA receptors strongly influences the ion channel function [13]. Depending on the neuronal tissue, the NMDA receptor subunits have different expression levels leading to a finetuning of NMDA receptor properties and contribution to different physiological processes like learning and cognitive function [14, 15]. Inhibition of NMDA receptor overstimulation by the open channel blocker memantine displays an important strategy for pharmacotherapy of moderate and severe AD that is characterized by pronounced loss of neuronal cells inducing excitotoxicity [16]. Since memantine is characterized as a weak and unselective open channel blocker its effectiveness is limited [16]. On the other hand, usage of more potent unselective channel blockers like ketamine is hampered by the severity of possible side effects [17]. Interestingly, several studies suggest that especially overstimulation of GluN2B containing NMDA receptors correlates with the observed neurodegeneration in AD or Huntington’s disease [6]. Thus, highly potent and GluN2B selective inhibitors might be promising drug candidates for better disease control and reduced side effects. The prototype of GluN2B specific NMDA receptor inhibitors is ifenprodil, which is used as a cerebral vasodilator due to its inhibitory effects on adrenergic receptors [18]. Structure reorganization of ifenprodil led to the development of WMS14-10 that shows increased potency at GluN2B containing NMDA receptors and an optimized selectivity profile towards adrenergic, serotonergic and sigma receptors in several different assays [19, 20]. To evaluate the capability of WMS14-10 to protect cells from excitotoxicity, a human neuronal cell model is needed to simulate excitotoxic conditions.
For this purpose, we used NMDA receptor expressing human SK-N-SH cells that were previously characterized as a cell model for neurodegenerative diseases like AD or Parkinson [21, 22]. SK-N-SH cells were isolated from bone marrow of a 4-year-old girl and exhibit characteristics of two neural progenitor cell types, a neuroblast type (N-type) and a substrate-adherent fibroblast-like or epithelial type (S-type) [23, 24]. It has been shown that upon all trans retinoic acid exposure, cell mitosis arrested and differentiation into a more neuronal phenotype takes place [25]. While NMDA receptors are also expressed on the progenitor cells, their expression level can be increased by neuronal differentiation [26]. Independently from the differentiation stage, the progenitor as well as the differentiated cells are sensitive to (S)-glutamate [26, 27]. However, NMDA-mediated cell death has only been assumed for the differentiated cell type [26].
In this project, we investigated the cell protective effect of the potent but unselective NMDA channel blocker ketamine as well as the GluN2B selective NMDA receptor inhibitor WMS14-10 against (S)-glutamate induced toxicity at the non-differentiated and differentiated SK-N-SH cells. Reverse transcriptase-PCR (RT-PCR) and immunostaining were performed for expression analysis of NMDA receptor subunits. Cell viability was analyzed in presence of (S)-glutamate and the NMDA receptor inhibitors by L-lactate dehydrogenase (LDH) and cell counting kit 8 (CCK8) assays as well as 4′,6-diamidino-2-phenylindole (DAPI) staining and subsequent flow cytometry. The results indicate that only a small fraction of (S)-glutamate induced toxicity is caused by NMDA receptor activation. Moreover, partial cell protection could only be achieved by unselective NMDA receptor inhibition, while GluN2B selective inhibition showed no effect. Our results indicate that neither non-differentiated nor differentiated SK-N-SH cells are a suitable cell model for the investigation of cell protective NMDA receptor inhibition.
Materials and Methods
TEVC measurements
Inhibitory activity of both compounds was evaluated via TEVC in Xenopus laevis oocytes (Ecocyte Bioscience, Dortmund, Germany) as previously described [19]. Harvesting of oocytes was carried out by Ecocyte Bioscience, which are certified by the Ministry of Nature, Environment and Consumer Protection NRW (Germany), in accordance with relevant guidelines and regulations. No further experiments with live vertebrates were performed. Oocytes were injected with 0.8 ng cRNA of GluN1-1a and 0.8 ng cRNA of GluN2B. The oocytes were incubated for 5 days in Barth`s solution, containing 88 mM NaCl, 1 mM KCl, 0.4 mM CaCl2, 0.33 mM Ca(NO3)2, 0.6 mM MgSO4, 5 mM TRIS-HCl, 2.4 mM NaHCO3, supplemented with 80 mg / L theophylline, 63 mg / L benzylpenicillin, 40 mg / L streptomycin, and 100 mg / L gentamycin at 18 °C. TEVC experiments were performed with Ba2+-Ringer solution (10 mM HEPES, 90 mM NaCl, 1 mM KCl, and 1.5 mM BaCl2, adjusted to pH 7.4). Agonist solution containing 5 to 50 mM (S)-glutamate and 0.4 mM glycine was freshly prepared from stock solutions in Ba2+-Ringer for each experiment. Recording was performed at -70 mV and recording pipettes (0.5 – 1.5 MΩ) were backfilled with 3 M KCl. Both compounds were tested at 5 - 10 independent oocytes from three different batches.
Cell culture
SK-N-SH cells (Merck, Darmstadt, Germany) were grown in Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Merck, Darmstadt, Germany) supplemented with 10 % FBS, 1x penicillin/streptomycin/glutamine (PSG) and 1x sodium pyruvate at 37 °C and 5 % CO2. The medium was changed every 3 days and 80 - 90 % confluent monolayers were passaged by 0.25 % Trypsin-EDTA (Merck). Cells were incubated for 3 min with Trypsin-EDTA, neutralized with medium and centrifuged for 3 min at 300 rpm.
Differentiation by RA
All-trans retinoic acid (RA) was dissolved in DMSO (10 mM). For differentiation, cells were plated into poly-L-ornithine-coated (0.01 %, Merck) wells at a density of 4.5 x 104 cells / cm2 and incubated for 24 h in the previously described media. To increase the sensitivity, cell number was increased to 3 x 105 cells / cm2 for intracellular Ca2+ recordings. Subsequently, the medium was changed to the differentiation medium (DMEM + 2.5 % FBS + 1x PSG + 10 µM RA, according to literature [28]). Cells were incubated with RA for 14 days with exchange of media every 48 h.
Reverse transcription and PCR
Total RNA was extracted using the NucleoSpin RNA Kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions. Reverse transcription (RT) was performed using the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher, USA) according to the procedure given by the manufacturer with 300 ng of total RNA used as template. PCR was performed in a Thermocycler (Eppendorf, Germany) using KOD hot start polymerase kit (Merck, Germany), 2 µL of cDNA from previous RT reaction and 0.6 µM of specific primers for NMDA receptor subunits (Table 1) [29]. Thermocycling conditions were chosen following the manufactures protocol. PCR products were evaluated on a 2 % RedSafe-stained agarose gel.

Table 1: Primer sequences
Immunostaining
For microscopy and immunostaining, cells were seeded on glass coverslips (VWR), which were plasma-treated for 1 min at 50 kV and subsequently coated with 0.001 % poly-L-ornithine in PBS for 60 min. The cells were then seeded onto the coverslip and incubated overnight in the media described above. First, the cells were washed twice with PBS and fixed with paraformaldehyde (4 % in PBS) for 30 min. Then, non-specific protein binding sites were saturated with 0.5 % bovine serum albumin in PBS for 60 min. Subsequently, primary antibodies (Table 2, 1:400 in 0.5% BSA/PBS) were added to the fixed cells and incubated overnight at 4 °C. On the following day, primary antibody solution was removed and cells were washed three times with PBS. Secondary antibodies (anti-Mouse HRP, GE Healthcare #NA931V, 1:800 in 0.5 % BSA/ PBS) and anti-Rabbit Alexa Fluor 488 (Sigma Aldrich #SAB4600036, 1:800 in 0.5 % BSA/ PBS) were added to the cells. Incubation was performed for 60 min under light exclusion at room temperature. After this, cells were washed again two times with PBS and one time with distilled water. In the first of the three washing steps, DAPI (Thermo Fisher #62247) 1:4000 was added to the PBS. The coverslip was then mounted on a slide using AquaPolymount (Polyscience) and was left hardening overnight. Coverslips were imaged using a confocal microscope (Leica DMI 4000 B) at a magnification of 63× (water immersion). Fluorescence intensity was analyzed using ImageJ 1.53t (NIH, USA) for every cell (n = 10-23) in every condition.
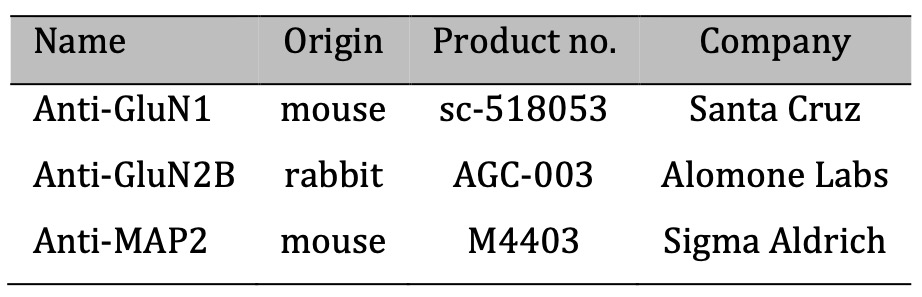
Table 2: Primer antibodies
Cell-counting Kit 8 for cell viability
Cell viability was evaluated using Cell-Counting Kit 8 (CCK-8) (Dojindo Laboratories, Japan) according to the manufacturers protocol. Cells were seeded into 96-well plates (1.5 x 104 cells / well) and incubated overnight at 37 °C and 5 % CO2. Media, containing different conditions, were changed afterwards. After 24 h of incubation, 10 µL of CCK-8 were applied to each well, the number of viable cells was evaluated after 3 h incubation time at 450 nm using an Ao absorbance microplate reader (Azure Biosystems, USA).
Lactate dehydrogenase assay for cell toxicity
To determine the cell toxicity of different test solutions, the lactate dehydrogenase (LDH) assay was used. In order to eliminate background absorption caused by the phenol red dye contained in normal DMEM (absorption maximum at pH 7.5: 560 nm), colorless DMEM (Sigma) was used for all subsequent steps. In addition, the concentration of FBS was lowered from 10 % to 5 %, since FBS already contains LDH at low concentrations and leads to background absorption. Subsequently, 1x PSG and 1x sodium pyruvate were added. This medium will be referred to as medium LDH in the following. To perform the cytotoxicity test for different substances, cells were seeded in 96-well plate and were incubated with medium LDH overnight. Subsequently, the cells were incubated for 24 h with the substances to be tested. About 45 min before the end of incubation, an additional triplet with 10 µL of water to monitor spontaneous activity and a triplet with 10 µL of cell lysis buffer to monitor maximum LDH activity were mixed. Next, 50 µL of the supernatant were transferred to a new cell culture plate and 50 µL of the reaction mixture were added and incubated for 30 min in the absence of light. After addition of the stop solution, the absorbance was measured at 492 nm and 680 nm for reference.
Annexin V/DAPI-staining and flow-cytometry
Annexin V/DAPI staining was used to analyze cell death via flow cytometry with a FACSAria III (BD Biosciences, USA). For FACS preparation, cells were incubated with the respective test substance for 24 h. After incubation, supernatant and detached cells were transferred to a falcon tube (15 mL). The cells were then centrifuged at 300 g for 3 min, resuspended in 500 µL of annexin V-binding buffer (140 mM NaCl, 2.5 mM CaCl2, 10 mM HEPES, pH = 7.4) and stored on ice. 50 µL of this cell suspension were mixed with 2.5 µL annexin V-AF568 and incubated for 10 min in the absence of light. Subsequently, 400 µL of annexin V-binding buffer containing DAPI at a ratio of 1:400 was added and incubated for 5 min. Cell suspension was then centrifuged at 300 g for 3 min, resuspended in 500 µL of annexin V-binding buffer and transferred to FACS tubes. 10, 000 events were recorded. Gating was set with the negative sample.
Bioelectrical impedance assay
To determine cell viability via bioelectrical impedance assay, the CardioExcyte96 (Nanion Technologies, Munich, Germany) was used. Methods and theoretical background were mentioned before [30–32]. Cells were seeded into 96-well plates (NSP-96 CardioExcyte96 Sensor Plates 2.0 mm, Nanion, Germany) (1.5x104 cells/well) and incubated overnight in the CardioExcyte96. Media, containing different conditions, were changed afterwards and cells were incubated for 24 hours. Cell growth was normalized to the signal at the time of application.
Intracellular Ca2+ recordings via Fluo-4-AM-based fluorescent plate reader analysis
Fluorescence based recordings were done as previously described [33, 34]. Cells were incubated with the fluorescent calcium indicator Fluo-4-AM at final concentration of 5 µM in the presence of Pluronic F127 (0.005 % w / v) and probenecid (3 mM) in Mg2+-free Krebs-Ringer solution (KRS, 121.0 mM NaCl, 2.5 mM KCl, 2.5 mM CaCl2, 1.0 mM NaH2PO4, 20.0 mM HEPES (pH 7.4), and 11.0 mM D-Glucose) for 60 min at 37°C. Subsequently the solution was changed to 150 µL of Mg2+-free KRS with 3 mM probenecid to prevent Fluo-4 leakage. Fluorescence was evaluated in a fluorescence plate reader (FLUOstar Omega, BMG Labtech, Germany) at 30 °C with an excitation wavelength of 480 nm and an emission wavelength of 520 nm with bottom optics. Fluorescence was recorded before and after application of 16.6 µL (1:10 dilution) of buffer, 100 mM KCl as a control or 50 mM (S)-glutamate [35, 36]. Changes in fluorescence are shown as ∆F/F0 (%), with ∆F as change of the fluorescence relative to the mean basal fluorescence (F0) before application of ligands, normalized to the maximum peak of ionomycin as control.
Statistical analysis and reproducibility
All values are given as mean ± SEM. Numbers of independent oocytes, analyzed cells or experiments are given in the main article, the figure legends or supplementary information with at least 3 independent replicates. Wherever applicable, data were statistical evaluated by one-way-ANOVA followed by post hoc mean comparison Tukey test or two-sited T-test using OriginPro 2024. P values are indicated by ns (not significant) for p > 0.05, * for p < 0.05, ** for p < 0.01 and *** for p < 0.001.
Results
(Non-)differentiated SK-N-SH cells express NMDA receptors
The SK-N-SH cell line is characterized by two different cell subtypes, the substrate-adherent type (S-type), recognizable by its flat and larger shape, and the neuronal type (N-type). The N-type has a distinctly different geometry, a thicker soma, as well as numerous neurite-like extensions [24]. The proportion of N-type cells can be increased by treatment with all-trans retinoic acid for 14 days [26]. Both conditions (non-differentiated / differentiated) were previously used for cellular disease modelling [37, 38].
Within 14 days of all-trans retinoic acid treatment an increase in the proportion of N-type cells as well as cell-cell connections between them can be observed (Fig. 1a, 1b). To verify the neuronal character of the N-type cells immunostaining against MAP2 (microtubule-associated protein 2) was conducted, which is typically expressed in dendritic structures of neuronal cells [39–41]. For SK-N-SH cells, MAP2 expression can be detected independently from the differentiation status (Fig. 1a, 1b). To utilize SK-N-SH cells as a cell model for NMDA receptor induced neurodegeneration, the cells do not only need neuronal properties but also high expression rates of functional NMDA receptors. Of particular interest are GluN2B containing NMDA receptors due to their functional association with neurodegenerative diseases [6]. Therefore, we performed whole-cell RNA isolation, reverse transcription followed by PCR to detect cDNA encoding for GluN1 and GluN2B subunits. The results indicate that mRNAs of both subunits are expressed independently from the maturation (Fig. 1c). In line with these results, co-immunostaining using GluN1 and GluN2B antibodies show that both receptor forming subunits are expressed at the plasma membrane (Fig. 1d, 1e). For differentiated cells expression of both subunits is also present at the cell-cell contacts (Fig. 1e). Interestingly, while fluorescence intensity for GluN1 subunit is not different for differentiated cells, fluorescence intensity of GluN2B subunit significantly rises in differentiated cells (SI Fig. 1). To generate functional NMDA signaling, the plasma membrane located subunits need to colocalize. Therefore, we analyzed the fraction of colocalized subunits using the fluorescence signal generated from immunostaining of GluN1 and GluN2B (Fig. 1f-1h) by Pearson’s coefficient. For non-differentiated cells, GluN1 and GluN2B fluorescence colocalized for 87 ± 2 % (n = 3), while the values for maturated cells were slightly reduced to 76 ± 4 % (n = 3). In summary, neuronal-like cells with colocalized GluN1 and GluN2B expressions were identified under non-differentiated and differentiated conditions rendering the expression of functional NMDA receptors as plausible for both conditions.
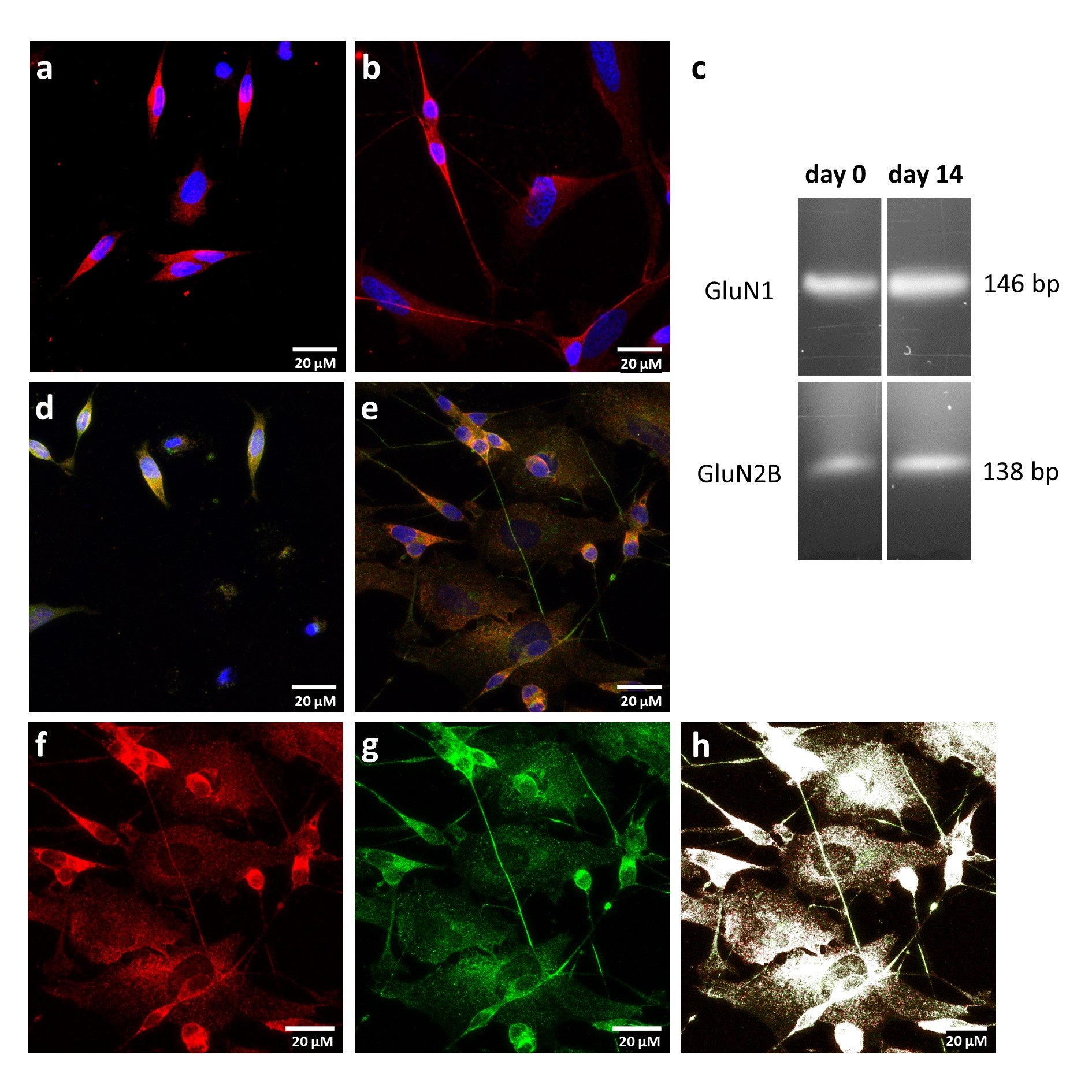
Fig. 1: Expression of NMDA receptor subunits. a, b Immunostaining of non-differentiated (a) and differentiated (b) SK-N-SH cells using MAP2 antibody (red) and DAPI (blue) c Cropped gel of RT-PCR products from whole cell RNA isolation using GluN1 and GluN2B primer. Non-cropped gel is listed as SI Figure 6 in the Supplementary Information. d, e Immunostaining of non-differentiated (d) and differentiated (e) SK-N-SH cells using GluN1 (red), GluN2B (green) antibodies and DAPI (blue). f-h Immunostaining of differentiated SK-N-SH cells showing GluN1 in red (f), GluN2B in green (g) and colocalized subunits in white (h).”
(S)-glutamate shows cytotoxic effects
Late stage neurodegeneration in AD is caused by elevation of extracellular (S)-glutamate concentrations resulting from cell disruptions subsequently leading to NMDA receptor overstimulation and cellular Ca2+ overload [42, 43]. To simulate these cytotoxic conditions at non-differentiated and differentiated SK-N-SH cells, different concentrations of (S)-glutamate were added to the cell media (DMEM high glucose) that also contains the co-agonist glycine (400 µM), which is required for effective NMDA receptor stimulation. After 24 h incubation time, cell viability was assessed by three different assays: cell counting kit 8 (CCK8), lactate dehydrogenase (LDH) and 4′,6-diamidin-2-phenylindol (DAPI) staining assay. The CCK8 assay uses a tetrazolium salt to detect overall dehydrogenase activity via colorimetric determination, while the LDH assay monitors the activity of lactate dehydrogenase that is released from non-viable cells. Additionally, ruptured cells were quantified by DAPI staining followed by fluorescence-activated cell sorting (FACS). The results summarized in Fig. 2a-2f and SI Figures 2-4 show that all assays detected significant reduction in cell viability under the influence of 50 mM (S)-glutamate for non-differentiated as well as for differentiated cells. Furthermore, reduced cell viability can also be observed under the influence of 25 mM (S)-glutamate for non-differentiated cells in CCK8 as well as DAPI/FACS assay (Fig. 2a, 2e). For differentiated cells, significant reduction of cell viability in presence of 25 mM (S)-glutamate was only detected via the LDH assay (Fig. 2d). Lower concentrations of (S)-glutamate did not significantly reduce SK-N-SH cell viability. These results indicate that non-differentiated and differentiated cells are sensitive to high concentrations of (S)-glutamate. Since only 50 mM (S)-glutamate led to a consistent and significant reduction in cell viability in all three assays, all further experiments were conducted using this concentration to simulate neurodegenerative conditions.
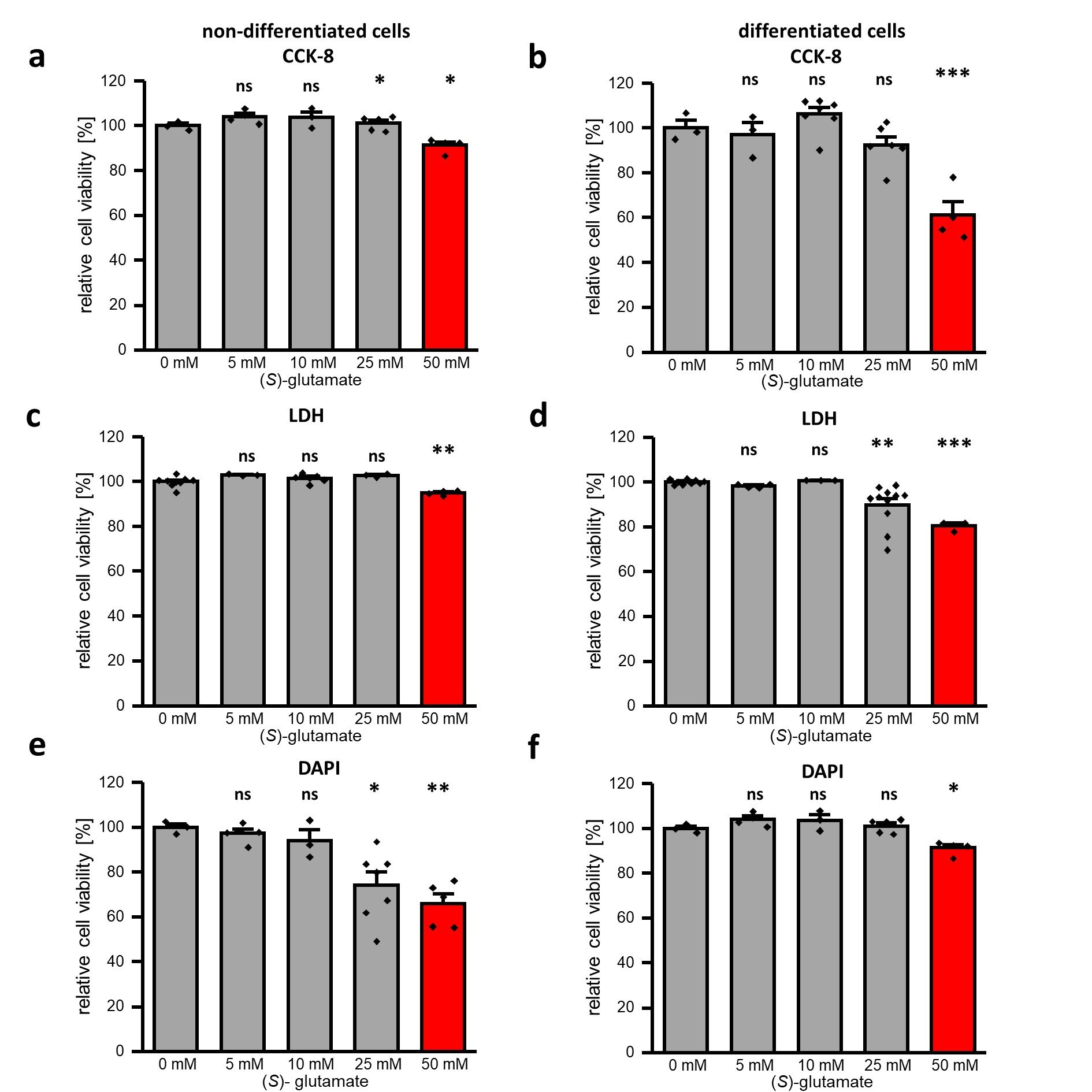
Fig. 2: Cell viability of (non-)differentiated SK-N-SH cells in presence of (S)-glutamate. Relative cell viability in presence of different (S)-glutamate concentrations was analyzed for non-differentiated (a, c, e) and differentiated SK-N-SH cells using CCK-8 (a, b), LDH (c, d) as well as DAPI/FACS (e, f) assays. Data were statistical evaluated by one-way-ANOVA followed by post hoc mean comparison Tukey test. P-values are indicated by ns (not significant) for p > 0.05, * for p<0.05, ** for p<0.01 and *** for p<0.001
Ketamine and WMS14-10 inhibit NMDA receptors at high (S)-glutamate concentrations
To evaluate the neuroprotective effect of unselective as well as GluN2B selective NMDA receptor inhibition, ketamine and WMS14-10 were utilized. Previous recordings via Two-electrode voltage clamp (TEVC) on GluN1/GluN2B expressing Xenopus laevis oocytes determined an IC50 value of 0.85 ± 0.25 µM for ketamine (Fig. 3a) in presence of 100 µM glycine and 100 µM (S)-glutamate [44]. For the GluN2B selective NMDA inhibitor rac-WMS14-10 (Fig. 3b), an IC50 value of 0.12 ± 0.01 µM at GluN1/GluN2B expressing oocytes was determined in presence of only 10 µM (S)-glutamate and 10 µM glycine, while no significant inhibition could be detected at GluN1/2A-, GluN1/2C- and GluN1/2D-containing NMDA receptors [19, 45]. Previous studies showed that some NMDA receptor modulators have an agonist-dependent activity that can be diminished or even abolished by high agonist concentrations [46–48]. To ensure that the inhibitory effect of ketamine as well as WMS14-10 is not influenced by high (S)-glutamate concentrations, we evaluated the inhibitory activity of 100 µM ketamine and 10 µM WMS14-10 at GluN1/GluN2B expressing oocytes in presence of 5 – 50 mM (S)-glutamate and 400 µM glycine by TEVC (Fig. 3c-3f). The results show that both compounds were able to generate almost full receptor inhibition at all tested (S)-glutamate levels (Fig. 3e, 3f). For ketamine current inhibition ranged from 92 ± 4 to 97 ± 8 % for the different (S)-glutamate levels, while WMS14-10 achieved inhibition values of 92 ± 7 to 99 ± 5 %. The mean values of inhibition for ketamine as well as WMS14-10 were not significantly different indicating no negative influence on the inhibitory effect by the used (S)-glutamate concentrations. Therefore, both compounds tender suitable to prevent NMDA receptor induced neurotoxicity at 50 mM (S)-glutamate in the SK-N-SH cell model.
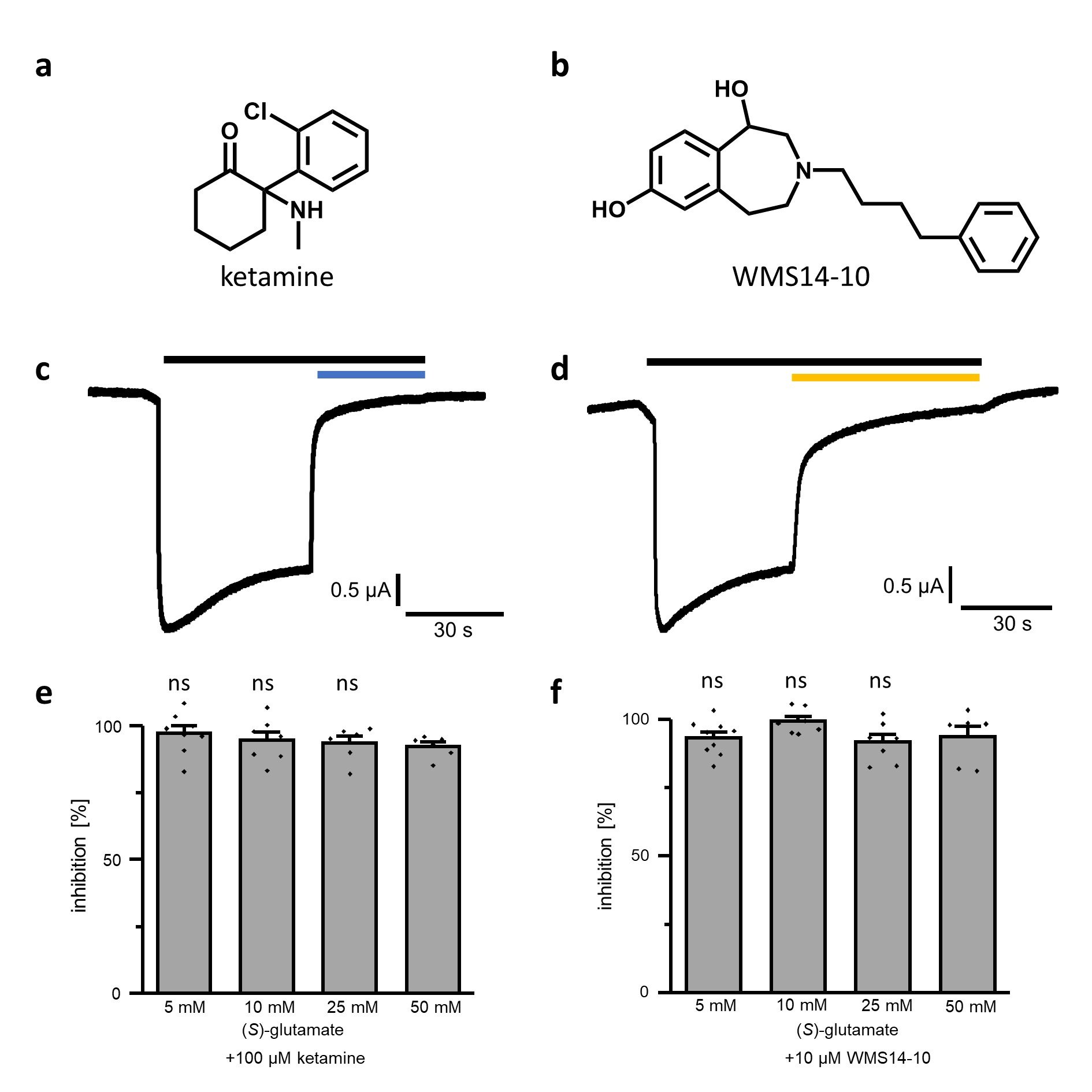
Fig. 3: Inhibitory activity at GluN1-1a/GluN2B expressing Xenopus laevis oocytes. a, b Structures of ketamine (a) and WMS14-10 (b). c, d Representative current traces of GluN1-1a/GluN2B expressing oocyte activated by 50 mM (S)-glutamate and 400 µM glycine (black bar), inhibited by 100 µM ketamine (c, blue bar) or 10 µM WMS14-10 (d, orange bar). e, f Inhibitory effect of 100 µM ketamine (e) and 10 µM WMS14-10 (f) in presence of different (S)-glutamate concentrations. Significance of mean differences was evaluated by one-way ANOVA in comparison to inhibitory effect in presence of 50 mM (S)-glutamate and is indicated by ns for p > 0.05
Ketamine shows a reversing effect on differentiated SK-N-SH cells
Before the cytoprotective potential of both compounds was evaluated under neurodegenerative conditions, off target toxicity at the SK-N-SH cells induced by 100 µM ketamine or 10 µM WMS14-10 needed to be excluded. Therefore, both compounds were applied to non-differentiated and differentiated cells and incubated for 24 h without further stimulation of (S)-glutamate (Fig. 4a, 4b). For both compounds no significant reduction of cell viability was observed in CCK8, LDH as well as in DAPI/FACS assay. Next, we evaluated the cytoprotective potential of both compounds in presence of 50 mM (S)-glutamate for 24 h (Fig. 4c, 4d). For non-differentiated cells, neither ketamine nor WMS14-10 were capable to significantly increase the cell viability in CCK8, LDH or DAPI/FACS assay compared to non-protected cells (Fig. 4c). Surprisingly, WMS14-10 even significantly reduced cell viability in the CCK8 assay for non-differentiated cells. A similar tendency was also observed in the CCK8 results for WMS14-10 without (S)-glutamate, although reduction of cell viability was not significant (Fig. 4a). In contrast to non-differentiated cells, significant cell protection was detected for 100 µM ketamine in CCK8 as well as in LDH assay. For DAPI/FACS slightly improved cell viability was observed. However, results were not significantly different to the control. In contrast to ketamine, subunit specific inhibition via WMS14-10 showed no protective effects in CCK8, LDH or DAPI/FACS assays. Similar to non-differentiated cells, WMS14-10 showed a tendency to reduce cell viability in CCK8 assay although the effect was not significant. In summary, cytoprotective effects were obtained by application of 100 µM ketamine for differentiated cells, while subunit specific inhibition via WMS14-10 did not improve cell survival.
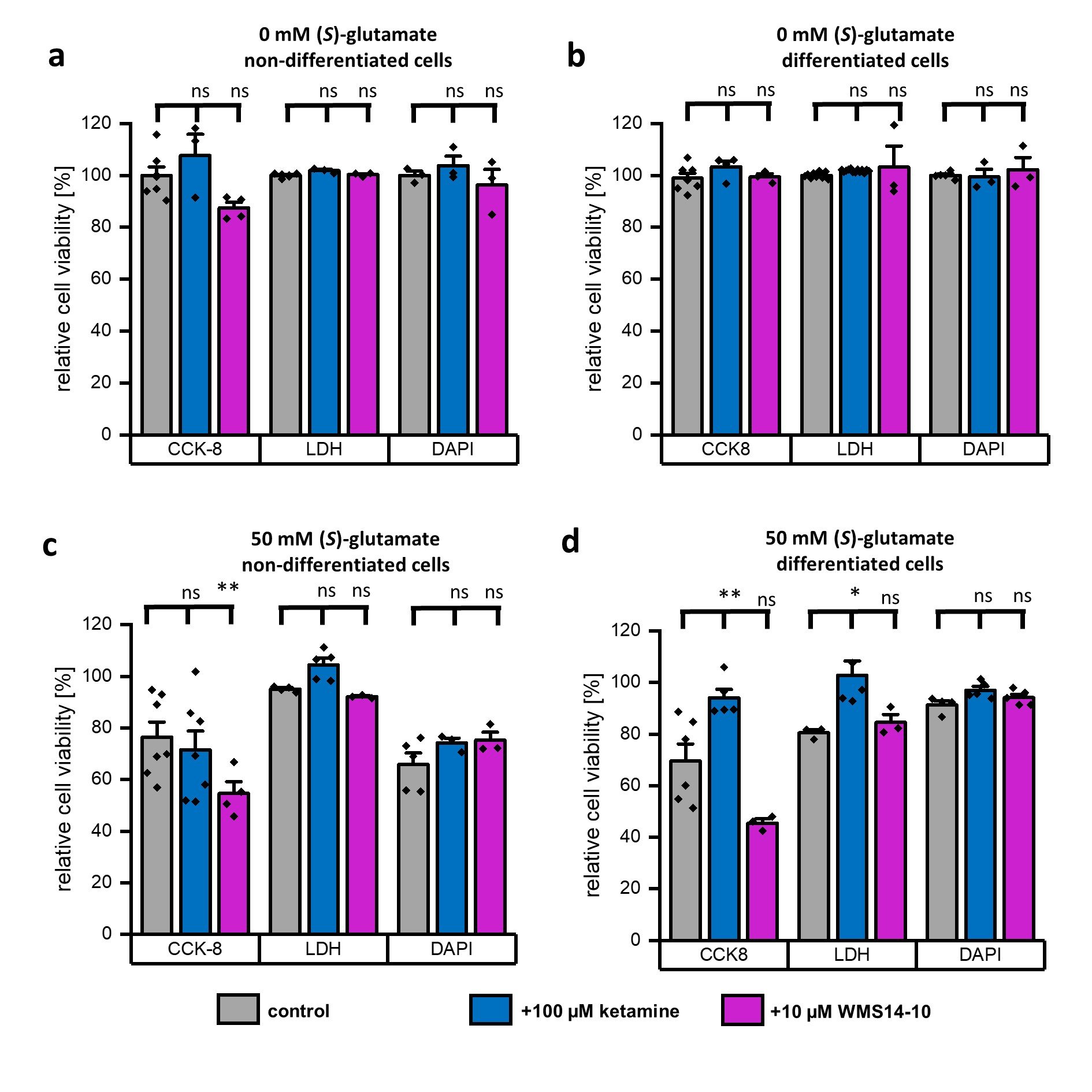
Fig. 4: Evaluation of cytoprotection via application of NMDA receptor inhibitors. a, b Relative cell viability of non-differentiated (a) and differentiated (b) SK-N-SH cells in presence of 100 µM ketamine (blue) or 10 µM WMS14-10 (magenta) without (S)-glutamate stimulation. Untreated cells (gray) serve as a control. c, d Relative cell viability of non-differentiated (c) and differentiated (d) SK-N-SH cells in presence of 50 mM (S)-glutamate without any NMDA receptor inhibitor (gray) as well as together with 100 µM ketamine (blue) or 10 µM WMS14-10 (magenta). Data were statistical evaluated by one-way-ANOVA followed by post hoc mean comparison Tukey test. P-values are indicated by ns (not significant) for p > 0.05, * for p<0.05, and ** for p<0.01
(S)-glutamate toxicity is immediately induced and not time-dependent
Previous cell viability experiments consistently show, that cytoprotective effects after 24 h are only achieved for differentiated cells in presence of 100 µM ketamine. However, even with this high concentration of ketamine full cell protection was not achieved even though similar ketamine concentrations were used in other NMDA receptor cell models [49–51]. These observations raised the question if the observed cell toxicity induced by (S)-glutamate is caused progressively over the 24 h incubation period. Previously performed assays (CCK8, LDH, DAPI/FACS) are only qualified to quantify the cell viability at the incubation endpoint. In order to analyze the time dependent neurodegeneration, we repeated the neurodegenerative experiments utilizing a bioelectrical impedance assay (Fig. 5). For this purpose, cells were plated on 96-well plates containing a reference and a sensing electrode (Fig. 5a). Without cells, current flow through the cell media is unimpeded (Fig. 5b), while cell growth leads to hindered current flow increasing the impedance (Fig. 5c). Under neurodegenerative conditions, detachment of cells causes impedance decrement that can be utilized to time-dependently quantify cell survival. Non-differentiated and differentiated SK-N-SH cells were seeded on 96 well plates with electrodes for impedance recordings and grown until maximum cell confluence indicated by constant impedance was reached (Fig. 5d, 5e, time index -8 to 0 h). Afterwards, 50 mM (S)-glutamate together with 100 µM ketamine or 10 µM WMS14-10 were added to the media (Fig. 5d, 5e, time index 0 h). As a control, the effect of 50 mM (S)-glutamate without any NMDA inhibitor as well as the cell growth without any (S)-glutamate application was analyzed. The results summarized in Fig. 5d to 5g show that application of 50 mM (S)-glutamate results in an instant and significant (p < 0.001) reduction of attached cells independently from the differentiation status as well as from co-application of NMDA receptor inhibitors ketamine and WMS14-10. Furthermore, application of (S)-glutamate almost abolished further cell growth within the observation time of 24 hours which is indicated by nearly constant growth curves (Fig. 5d, 5e). In contrast, non-differentiated and differentiated cells that were not exposed to (S)-glutamate show constant growth within the 24 h (Fig. 5d, 5e). Evaluation after 24 h illustrates that the most prominent reduction of cell growth for non-differentiated and differentiated cells can be detected under the influence of 50 mM (S)-glutamate without application of any NMDA receptor inhibitor (Fig. 5f, 5g). Co-application of neither 100 µM ketamine nor 10 µM WMS14-10 was able to significantly improve the cell survival. However, for differentiated cells, application of 100 µM ketamine shows a tendency for cell protection by increasing the growth value from -50.6 ± 4.9 % (n = 10) for the unprotected condition to -36.6 ± 1.5 SEM (n = 8).
This observation is in line with the previous results and might indicate an increase of functional NMDA receptor expression for differentiated cells, which should result in an increased Ca2+ influx by (S)-glutamate application. To investigate this hypothesis in more detail, we performed intracellular Ca2+ recordings using the fluorescence calcium indicator Fluo-4-AM (SI Fig. 5). Application of 100 mM K+ as well as terminal application of ionomycin to each condition served as a positive control. Independently from the differentiation status, application of 50 mM (S)-glutamate failed to induce significantly increased Ca2+ signals compared to non-treated cells even though a slightly increased fluorescence signal can be observed for differentiated cells (SI Fig. 5c, 5d). In summary, impedance as well as intracellular Ca2+ recordings indicate that (S)-glutamate induced toxicity is immediately occurring after application. Due to the almost unchanged toxicity under presence of NMDA receptor inhibitors and the low Ca2+ signals it can be concluded that only a small part of the induced toxicity is caused by overstimulation of NMDA receptors.
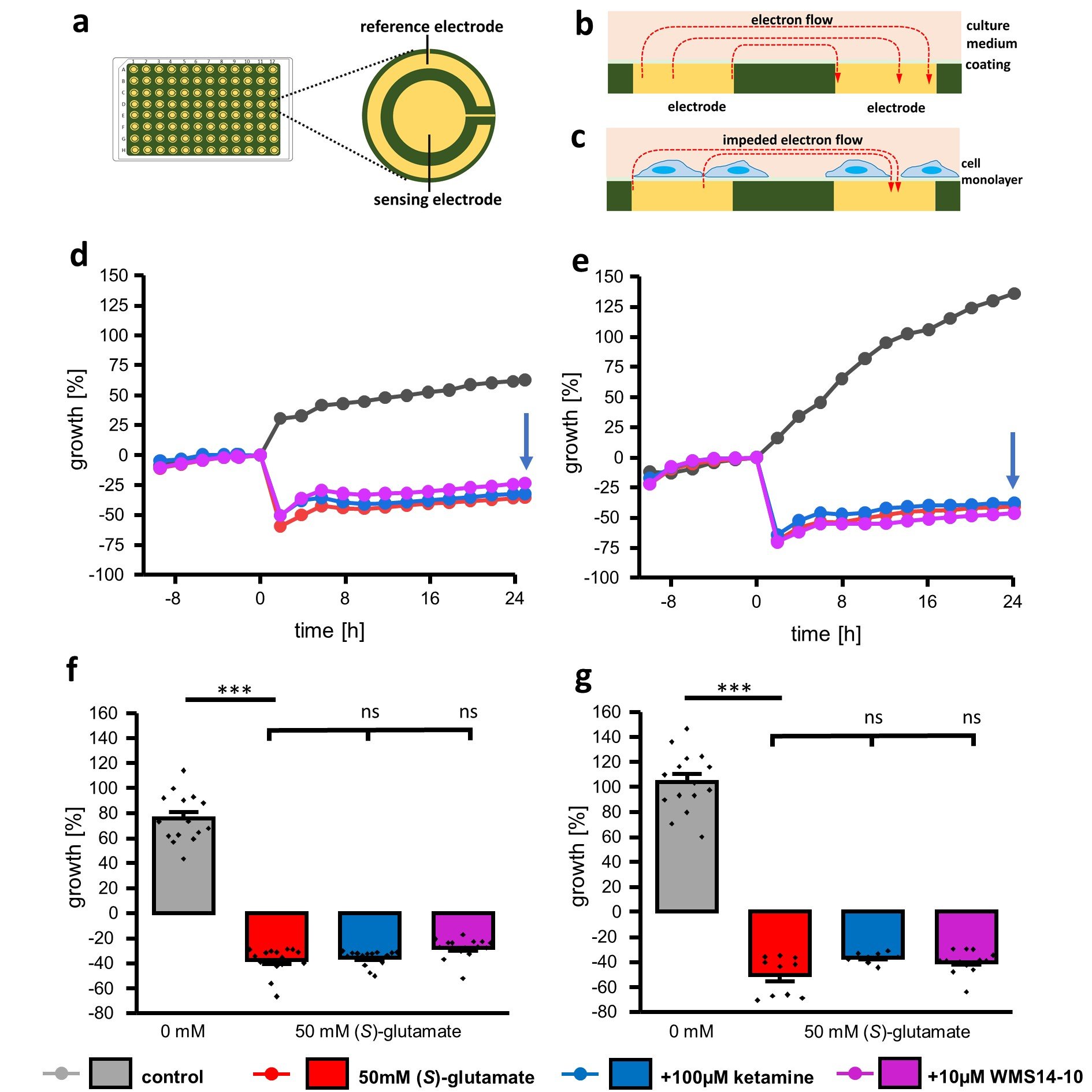
Fig. 5: Time-dependent cytotoxicity evaluated via bioelectrical impedance assay. a Illustration of the 96-well plate and a single well with reference and sensing electrode. b, c Side-view of a single well without (b) and with (c) attached cells leading to an alteration of impedance recordings. d, e Representative proliferation graphs of non-differentiated (d) and differentiated (e) cells incubated with 0 mM (S)-glutamate (gray), 50 mM (S)-glutamate without NMDA receptor inhibitor (red) as well as 50 mM (S)-glutamate together with 100 µM ketamine (blue) or 10 µM WMS14-10 (magenta). Application of different compounds was performed at timepoint 0 h. f, g Growth evaluation of non-differentiated (f) and differentiated (g) cells 24 h after application of different compounds (blue arrow in d, e). Data were statistical evaluated by one-way-ANOVA followed by post hoc mean comparison Tukey test. P-values are indicated by ns (not significant) for p > 0.05 and *** for p<0.001
Discussion
Within this study, we analyzed the suitability of non-differentiated and differentiated SK-N-SH cells as a human cell model for neurodegeneration induced by NMDA receptor overstimulation. Of particular interest is the functional expression of GluN2B containing NMDA receptors due to their role in different neurodegenerative diseases.
In line with previous studies, we showed expression and colocalization of the NMDA receptor forming subunits GluN1 and GluN2B in both non-differentiated and differentiated SK-N-SH cells, which is needed for functional NMDA receptor formation [26]. However, in comparison to these results an increase in fluorescence intensity was only detected for the GluN2B subunit in differentiated cells. Furthermore, we observed a neuronal phenotype including expression of the neuronal marker MAP2 under both conditions as previously described [24, 25].
Previous studies lacking a direct functional comparison between non-differentiated and differentiated cells in different assays hampering the critical appraisal how the differentiation status influences the suitability of SK-N-SH cells to serve as a NMDA receptor cell model [25, 26, 28, 38, 52]. Our results show that the presence of more than 25 mM (S)-glutamate lead to a reduction in cell viability, which is almost independent from the differentiation status. This concentration is at least 1000-fold higher than the EC50 value of (S)-glutamate activating GluN1/GluN2B containing NMDA receptors expressed in Xenopus laevis oocytes [53]. Moreover, this concentration is also several fold higher than the (S)-glutamate concentrations used in other NMDA receptor cell models indicating that the reduction in cell viability might be not exclusively induced by NMDA receptor overactivation but also overstimulation of other glutamatergic receptors [49, 54, 55].
Beside the induction of non-NMDA receptor mediated effects high agonist concentrations can also influence the activity of NMDA receptor inhibitors [46–48]. Therefore, the inhibitory effect of ketamine and WMS14-10 was analyzed in presence of high (S)-glutamate concentrations in Xenopus laevis oocytes. The results show that both compounds are able to fully inhibit the NMDA receptor even in presence of high (S)-glutamate concentrations without any significant loss in compound effectiveness. Consequently, both compounds should prevent NMDA receptor overactivation during neurodegenerative conditions. Simultaneously, supplementation of SK-N-SH cell media with 100 µM ketamine or 10 µM WMS14-10 for 24 h did not cause any significant reduction in cell viability excluding severe compound induced toxicity.
In presence of 50 mM (S)-glutamate, partial cell protection was only achieved by 100 µM ketamine using differentiated SK-N-SH cells, while 10 µM WMS14-10 failed to protect both undifferentiated and differentiated cells, even though the expression of GluN2B subunit rises in differentiated cells as indicated by fluorescence intensity. With respect to the previous data for 10 µM WMS14-10 these results indicate that the (S)-glutamate induced toxicity is not primarily generated by overactivation of GluN2B containing NMDA receptors. On the contrary, cell protection by 100 µM ketamine using differentiated SK-N-SH cells suggest a partial contribution of NMDA receptors to the induced cell death. However, main toxicity is not generated by NMDA receptor overactivation as visualized by the bioimpedance recordings. Furthermore, the protective effect of 100 µM ketamine might be also induced by indirect off-target effects due to residual activity at α7, Cav1.2 or high affinity state of the D2 receptors [56–58].
The minor protective effect of ketamine as an unselective NMDA channel blocker in differentiated cells is also in accordance with the low intracellular Ca2+ signals. Although our plate-based assay showed a minimal rise of Ca2+ levels in differentiated cells compared to native cells under the influence of 50 mM (S)-glutamate, the overall Ca2+ signal was not significantly different to Ca2+ levels in untreated cells. In line with this, previous single cell recordings could not detect any Ca2+ signals in non-differentiated cells under NMDA stimulating conditions [26].
Conclusion
In summary, our results consistently show that neither native nor differentiated SK-N-SH cells are suitable to serve as a neurodegenerative cell model for NMDA receptor induced excitotoxicity. While (S)-glutamate toxicity in native SK-N-SH cells is completely independent from the NMDA receptor, cytotoxic effects in differentiated cells depend only to a minor extend on the overstimulation of NMDA receptors hampering a clear identification of novel neuroprotective NMDA receptor inhibitors. Furthermore, this small fraction of NMDA receptor-induced toxicity is not exclusively mediated by GluN2B containing NMDA receptors since only ketamine as an unselective inhibitor was able to partially inhibit cell toxicity.
Acknowledgements
The study was founded by the Innovative Medizinische Forschung (IMF SC-112107) Münster, who is gratefully acknowledged, and conceptualized by GG and JAS. Experiments were performed by GG supported by PD, SP, NR and CB. The manuscript was written by GG and JAS supported by NSS and GS. All authors critically revised the final manuscript. We thank Nanion for giving us the opportunity to use their CardioExcyte 96. Furthermore, we thank Prof. Dr. Bernhard Wünsch for providing WMS14-10, Prof. Dr. Frank Rosenbauer as well as Thorsten König for FACS analyses and Anne Humberg for overall support. The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M: Global prevalence of dementia: a Delphi consensus study. Lancet 2005;366:2112-7.
https://doi.org/10.1016/S0140-6736(05)67889-0 |
| 2 | Ou Z, Pan J, Tang S, Duan D, Yu D, Nong H, Wang Z: Global Trends in the Incidence, Prevalence, and Years Lived With Disability of Parkinson's Disease in 204 Countries/Territories From 1990 to 2019. Front Public Health 2021;9:776847.
https://doi.org/10.3389/fpubh.2021.776847 |
| 3 | Medina A, Mahjoub Y, Shaver L, Pringsheim T: Prevalence and Incidence of Huntington's Disease: An Updated Systematic Review and Meta-Analysis. Mov Disord 2022;37:2327-35.
https://doi.org/10.1002/mds.29228 |
| 4 | Olney JW, Wozniak DF, Farber NB: Excitotoxic neurodegeneration in Alzheimer disease. New hypothesis and new therapeutic strategies. Arch Neurol 1997;54:1234-40.
https://doi.org/10.1001/archneur.1997.00550220042012 |
| 5 | Zhang Y, Li P, Feng J, Wu M: Dysfunction of NMDA receptors in Alzheimer's disease. Neurol Sci 2016;37:1039-47.
https://doi.org/10.1007/s10072-016-2546-5 |
| 6 | Parsons MP, Raymond LA: Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron 2014;82:279-93.
https://doi.org/10.1016/j.neuron.2014.03.030 |
| 7 | Li L, Fan M, Icton CD, Chen N, Leavitt BR, Hayden MR, Murphy TH, Raymond LA: Role of NR2B-type NMDA receptors in selective neurodegeneration in Huntington disease. Neurobiol Aging 2003;24:1113-21.
https://doi.org/10.1016/j.neurobiolaging.2003.04.003 |
| 8 | Hasan MT, Hernández-González S, Dogbevia G, Treviño M, Bertocchi I, Gruart A, Delgado-García JM: Role of motor cortex NMDA receptors in learning-dependent synaptic plasticity of behaving mice. Nat Commun 2013;4:2258.
https://doi.org/10.1038/ncomms3258 |
| 9 | Wong JH, Muthuraju S, Reza F, Senik MH, Zhang J, Mohd Yusuf Yeo NAB, Chuang HG, Jaafar H, Yusof SR, Mohamad H, Tengku Muhammad TS, Ismail NH, Husin SS, Abdullah JM: Differential expression of entorhinal cortex and hippocampal subfields α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptors enhanced learning and memory of rats following administration of Centella asiatica. Biomed Pharmacother 2019;110:168-80.
https://doi.org/10.1016/j.biopha.2018.11.044 |
| 10 | Berlese DB, Sauzem PD, Carati MC, Guerra GP, Stiegemeier JA, Mello CF, Rubin MA: Time-dependent modulation of inhibitory avoidance memory by spermidine in rats. Neurobiol Learn Mem 2005;83:48-53.
https://doi.org/10.1016/j.nlm.2004.07.004 |
| 11 | Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH: Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 1992;256:1217-21.
https://doi.org/10.1126/science.256.5060.1217 |
| 12 | Ulbrich MH, Isacoff EY: Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A 2008;105:14163-8.
https://doi.org/10.1073/pnas.0802075105 |
| 13 | Wyllie DJA, Livesey MR, Hardingham GE: Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology 2013;74:4-17.
https://doi.org/10.1016/j.neuropharm.2013.01.016 |
| 14 | Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N: Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. . J Comp Neurol 1994;347:150-60.
https://doi.org/10.1002/cne.903470112 |
| 15 | Bashir ZI, Alford S, Davies SN, Randall AD, Collingridge GL: Long-term potentiation of NMDA receptor-mediated synaptic transmission in the hippocampus. Nature 1991;349:156-8.
https://doi.org/10.1038/349156a0 |
| 16 | Molinuevo JL, Lladó A, Rami L: Memantine: targeting glutamate excitotoxicity in Alzheimer's disease and other dementias. Am J Alzheimers Dis Other Demen 2005;20:77-85.
https://doi.org/10.1177/153331750502000206 |
| 17 | Mohammad Shehata I, Masood W, Nemr N, Anderson A, Bhusal K, Edinoff AN, Cornett EM, Kaye AM, Kaye AD: The Possible Application of Ketamine in the Treatment of Depression in Alzheimer's Disease. Neurol Int 2022;14:310-21.
https://doi.org/10.3390/neurolint14020025 |
| 18 | Borza I, Domány G: NR2B selective NMDA antagonists: the evolution of the ifenprodil-type pharmacophore. Curr Top Med Chem 2006;6:687-95.
https://doi.org/10.2174/156802606776894456 |
| 19 | Schreiber JA, Schepmann D, Frehland B, Thum S, Datunashvili M, Budde T, Hollmann M, Strutz-Seebohm N, Wünsch B, Seebohm G: A common mechanism allows selective targeting of GluN2B subunit-containing N-methyl-D-aspartate receptors. Commun Biol 2019;2:420.
https://doi.org/10.1038/s42003-019-0645-6 |
| 20 | Tewes B, Frehland B, Schepmann D, Schmidtke K-U, Winckler T, Wünsch B: Conformationally constrained NR2B selective NMDA receptor antagonists derived from ifenprodil: Synthesis and biological evaluation of tetrahydro-3-benzazepine-1, 7-diols. Bioorg Med Chem 2010;18:8005-15.
https://doi.org/10.1016/j.bmc.2010.09.026 |
| 21 | Green PS, Gridley KE, Simpkins JW: Estradiol protects against beta-amyloid (25-35)-induced toxicity in SK-N-SH human neuroblastoma cells. Neurosci Lett 1996;218:165-8.
https://doi.org/10.1016/S0304-3940(96)13148-7 |
| 22 | Belkacemi A, Ramassamy C: Innovative Anthocyanin/Anthocyanidin Formulation Protects SK-N-SH Cells Against the Amyloid-β Peptide-Induced Toxicity: Relevance to Alzheimer's Disease. Cent Nerv Syst Agents Med Chem 2015;16:37-49.
https://doi.org/10.2174/1871524915666150730125532 |
| 23 | Maeda K, Feyles V, McGarry RC, Jerry LM: Melanocytic differentiation of human neuroblastoma: expression of a human melanosome-associated antigen. J Invest Dermatol 1990;95:665-70.
https://doi.org/10.1111/1523-1747.ep12514336 |
| 24 | Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA: Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res 1989;49:219-25.
|
| 25 | Wainwright LJ, Lasorella A, Iavarone A: Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc Natl Acad Sci U S A 2001;98:9396-400.
https://doi.org/10.1073/pnas.161288698 |
| 26 | Pizzi M, Boroni F, Bianchetti A, Moraitis C, Sarnico I, Benarese M, Goffi F, Valerio A, Spano P: Expression of functional NR1/NR2B-type NMDA receptors in neuronally differentiated SK-N-SH human cell line. Eur J Neurosci 2002;16:2342-50.
https://doi.org/10.1046/j.1460-9568.2002.02403.x |
| 27 | Peng L, Zhang X, Cui X, Zhu D, Wu J, Sun D, Yue Q, Li Z, Liu H, Li G, Zhang J, Xu H, Liu F, Qin C, Li M, Sun J: Paliperidone protects SK-N-SH cells against glutamate toxicity via Akt1/GSK3β signaling pathway. Schizophr Res 2014;157:120-7.
https://doi.org/10.1016/j.schres.2014.05.037 |
| 28 | Cheung Y-T, Lau WK-W, Yu M-S, Lai CS-W, Yeung S-C, So K-F, Chang RC-C: Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology 2009;30:127-35.
https://doi.org/10.1016/j.neuro.2008.11.001 |
| 29 | Anaparti V, Pascoe CD, Jha A, Mahood TH, Ilarraza R, Unruh H, Moqbel R, Halayko AJ: Tumor necrosis factor regulates NMDA receptor-mediated airway smooth muscle contractile function and airway responsiveness. Am J Physiol Lung Cell Mol Physiol 2016;311:L467-80.
https://doi.org/10.1152/ajplung.00382.2015 |
| 30 | Arndt S, Seebach J, Psathaki K, Galla H-J, Wegener J: Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens Bioelectron 2004;19:583-94.
https://doi.org/10.1016/S0956-5663(03)00269-0 |
| 31 | Bot C, Guinot D, Thomas U, Doerr L, Stoelzle-Feix S, Beckler M, Okeyo G, Oestreich J, Haedo R, George M, Fertig N: Cardiotoxicity screening using the CardioExcyte 96: A noninvasive methodology of combining extracellular field potential and impedance measurements. J Pharmacol Toxicol Methods 2015;75:192-3.
https://doi.org/10.1016/j.vascn.2015.08.117 |
| 32 | Yin H, Wang* FL, Wang AL, Cheng J, Zhou Y: Bioelectrical Impedance Assay to Monitor Changes in Aspirin‐Treated Human Colon Cancer HT‐29 Cell Shape during Apoptosis. Anal Lett 2007;40:85-94.
https://doi.org/10.1080/00032710600952424 |
| 33 | Nguyen HN, Wang C, Perry DC: Depletion of intracellular calcium stores is toxic to SH-SY5Y neuronal cells. Brain Res 2002;924:159-66.
https://doi.org/10.1016/S0006-8993(01)03229-2 |
| 34 | Strünker T, Goodwin N, Brenker C, Kashikar ND, Weyand I, Seifert R, Kaupp UB: The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature 2011;471:382-6.
https://doi.org/10.1038/nature09769 |
| 35 | Tay B, Stewart TA, Davis FM, Deuis JR, Vetter I: Development of a high-throughput fluorescent no-wash sodium influx assay. PloS One 2019;14:e0213751.
https://doi.org/10.1371/journal.pone.0213751 |
| 36 | Gee KR, Brown KA, Chen WN, Bishop-Stewart J, Gray D, Johnson I: Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell Calcium 2000;27:97-106.
https://doi.org/10.1054/ceca.1999.0095 |
| 37 | Akkuratov EE, Wu J, Sowa D, Shah ZA, Liu L: Ouabain-Induced Signaling and Cell Survival in SK-N-SH Neuroblastoma Cells Differentiated by Retinoic Acid. CNS Neurol Disord Drug Targets 2015;14:1343-9.
https://doi.org/10.2174/1871527314666150821103008 |
| 38 | Chang P-A, Wu Y-J, Li W, Leng X-F: Effect of carbamate esters on neurite outgrowth in differentiating human SK-N-SH neuroblastoma cells. Chem Biol Interact 2006;159:65-72.
https://doi.org/10.1016/j.cbi.2005.09.005 |
| 39 | Izant JG, McIntosh JR: Microtubule-associated proteins: a monoclonal antibody to MAP2 binds to differentiated neurons. Proc Natl Acad Sci U S A 1980;77:4741-5.
https://doi.org/10.1073/pnas.77.8.4741 |
| 40 | Cassimeris L, Spittle C: Regulation of microtubule-associated proteins. Int Rev Cytol 2001;210:163-226.
https://doi.org/10.1016/S0074-7696(01)10006-9 |
| 41 | Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N: MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol 2002;158:541-9.
https://doi.org/10.1083/jcb.200110134 |
| 42 | McBain CJ, Mayer ML: N-methyl-D-aspartic acid receptor structure and function. Physiol Rev 1994;74:723-60.
https://doi.org/10.1152/physrev.1994.74.3.723 |
| 43 | Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, Traynelis SF: Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol 2018;150:1081-105.
https://doi.org/10.1085/jgp.201812032 |
| 44 | Zhang Y, Ye F, Zhang T, Lv S, Zhou L, Du D, Lin H, Guo F, Luo C, Zhu S: Structural basis of ketamine action on human NMDA receptors. Nature 2021;596:301-5.
https://doi.org/10.1038/s41586-021-03769-9 |
| 45 | Börgel F, Szermerski M, Schreiber JA, Temme L, Strutz-Seebohm N, Lehmkuhl K, Schepmann D, Ametamey SM, Seebohm G, Schmidt TJ, Wünsch B: Synthesis and Pharmacological Evaluation of Enantiomerically Pure GluN2B Selective NMDA Receptor Antagonists. ChemMedChem 2018;13:1580-7.
https://doi.org/10.1002/cmdc.201800214 |
| 46 | Schreiber JA, Müller SL, Westphälinger SE, Schepmann D, Strutz-Seebohm N, Seebohm G, Wünsch B: Systematic variation of the benzoylhydrazine moiety of the GluN2A selective NMDA receptor antagonist TCN-201. Eur J Med Chem 2018;158:259-69.
https://doi.org/10.1016/j.ejmech.2018.09.006 |
| 47 | Lind GE, Mou T-C, Tamborini L, Pomper MG, Micheli C de, Conti P, Pinto A, Hansen KB: Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits. Proc Natl Acad Sci U S A 2017;114:E6942-E6951.
https://doi.org/10.1073/pnas.1707752114 |
| 48 | Edman S, McKay S, Macdonald LJ, Samadi M, Livesey MR, Hardingham GE, Wyllie DJA: TCN 201 selectively blocks GluN2A-containing NMDARs in a GluN1 co-agonist dependent but non-competitive manner. Neuropharmacology 2012;63:441-9.
https://doi.org/10.1016/j.neuropharm.2012.04.027 |
| 49 | Temme L, Schepmann D, Schreiber JA, Frehland B, Wünsch B: Comparative Pharmacological Study of Common NMDA Receptor Open Channel Blockers Regarding Their Affinity and Functional Activity toward GluN2A and GluN2B NMDA Receptors. ChemMedChem 2018;13:446-52.
https://doi.org/10.1002/cmdc.201700810 |
| 50 | Boeckman FA, Aizenman E: Pharmacological properties of acquired excitotoxicity in Chinese hamster ovary cells transfected with N-methyl-D-aspartate receptor subunits. J Pharmacol Exp Ther 1996;279:515-23.
|
| 51 | Steinmetz RD, Fava E, Nicotera P, Steinhilber D: A simple cell line based in vitro test system for N-methyl-D-aspartate (NMDA) receptor ligands. J Neurosci Methods 2002;113:99-110.
https://doi.org/10.1016/S0165-0270(01)00482-4 |
| 52 | Mak YT, Lam WP, Lü L, Wong YW, Yew DT: The toxic effect of ketamine on SH-SY5Y neuroblastoma cell line and human neuron. Microsc Res Tech 2010;73:195-201.
https://doi.org/10.1002/jemt.20774 |
| 53 | Erreger K, Geballe MT, Kristensen A, Chen PE, Hansen KB, Lee CJ, Yuan H, Le P, Lyuboslavsky PN, Micale N, Jørgensen L, Clausen RP, Wyllie DJA, Snyder JP, Traynelis SF: Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol Pharmacol 2007;72:907-20.
https://doi.org/10.1124/mol.107.037333 |
| 54 | Sinor JD, Du S, Venneti S, Blitzblau RC, Leszkiewicz DN, Rosenberg PA, Aizenman E: NMDA and glutamate evoke excitotoxicity at distinct cellular locations in rat cortical neurons in vitro. J Neurosci 2000;20:8831-7.
https://doi.org/10.1523/JNEUROSCI.20-23-08831.2000 |
| 55 | Gupta K, Hardingham GE, Chandran S: NMDA receptor-dependent glutamate excitotoxicity in human embryonic stem cell-derived neurons. Neurosci Lett 2013;543:95-100.
https://doi.org/10.1016/j.neulet.2013.03.010 |
| 56 | Moaddel R, Abdrakhmanova G, Kozak J, Jozwiak K, Toll L, Jimenez L, Rosenberg A, Tran T, Xiao Y, Zarate CA, Wainer IW: Sub-anesthetic concentrations of (R,S)-ketamine metabolites inhibit acetylcholine-evoked currents in α7 nicotinic acetylcholine receptors. Eur J Pharmacol 2013;698:228-34.
https://doi.org/10.1016/j.ejphar.2012.11.023 |
| 57 | Kapur S, Seeman P: Ketamine has equal affinity for NMDA receptors and the high-affinity state of the dopamine D2 receptor. Biol Psychiatry 2001;49:954-7.
https://doi.org/10.1016/S0006-3223(01)01110-6 |
| 58 | Chen H, Vandorpe DH, Xie X, Alper SL, Zeidel ML, Yu W: Disruption of Cav1.2-mediated signaling is a pathway for ketamine-induced pathology. Nat Commun 2020;11:4328.
https://doi.org/10.1038/s41467-020-18167-4 |